1 # L2 normalization之实现:
2 # 1. 获取数据
3 # 2. 封装数据
4 # 3. 构建模型
5 # 4. 构建损失函数
6 # 5. 构建优化器
7 # 6. 训练单元
8 # 7. 测试单元
9 # 8. 启动训练与测试
10 # 9. 保存模型
11
12 import numpy
13 import torch
14 from torch import optim
15 from matplotlib import pyplot as plt
16
17
18 numpy.random.seed(0)
19 torch.random.manual_seed(0)
20
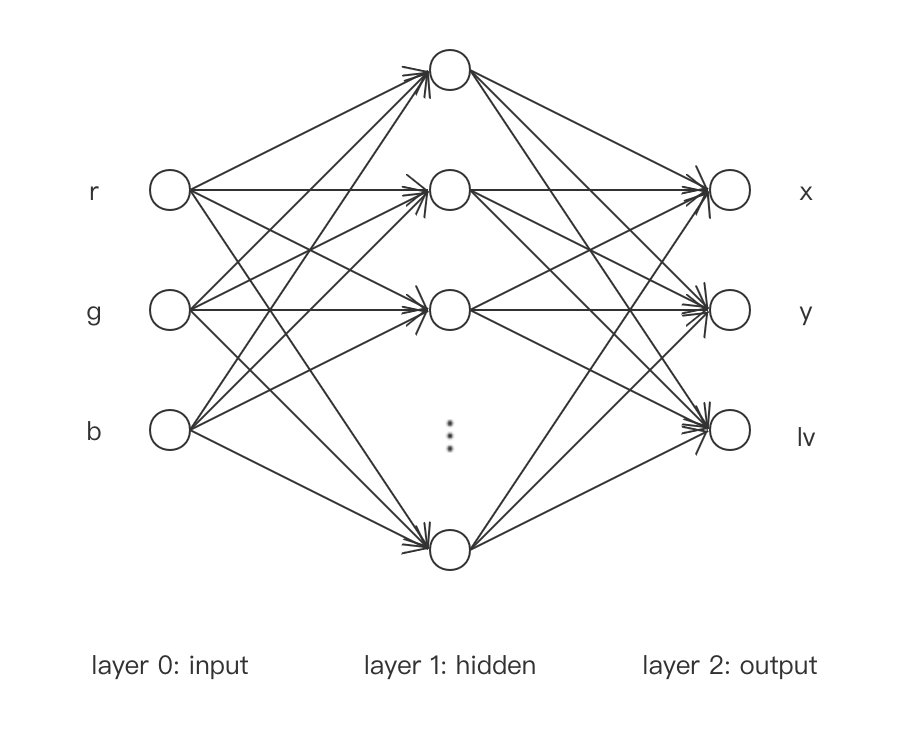
21 def xFunc(r, g, b):
22 x = r + 2 * g + 3 * b
23 return x
24
25 def yFunc(r, g, b):
26 y = r ** 2 + 2 * g ** 2 + 3 * b ** 2
27 return y
28
29 def lvFunc(r, g, b):
30 lv = -3 * r - 4 * g - 5 * b
31 return lv
32
33
34 # 1. 获取数据
35 class GeneData(object):
36
37 def __init__(self, rRange=[-1, 1], gRange=[-1, 1], bRange=[-1, 1]):
38 self.__rRange = rRange
39 self.__gRange = gRange
40 self.__bRange = bRange
41
42 def getDataset(self, num):
43 rArr, gArr, bArr = self.__generate_rgbArr(num)
44 xArr, yArr, lvArr = self.__generate_xylvArr(rArr, gArr, bArr)
45 rgb = numpy.hstack((rArr.reshape((-1, 1)), gArr.reshape((-1, 1)), bArr.reshape((-1, 1))))
46 xylv = numpy.hstack((xArr.reshape((-1, 1)), yArr.reshape((-1, 1)), lvArr.reshape((-1, 1))))
47 return torch.tensor(rgb, dtype=torch.float), torch.tensor(xylv, dtype=torch.float)
48
49 def __generate_xylvArr(self, rArr, gArr, bArr):
50 xArr = xFunc(rArr, gArr, bArr)
51 yArr = yFunc(rArr, gArr, bArr)
52 lvArr = lvFunc(rArr, gArr, bArr)
53 return xArr, yArr, lvArr
54
55 def __generate_rgbArr(self, num):
56 rArr = numpy.random.uniform(*self.__rRange, num)
57 gArr = numpy.random.uniform(*self.__gRange, num)
58 bArr = numpy.random.uniform(*self.__bRange, num)
59 return rArr, gArr, bArr
60
61
62 # 2. 封装数据
63 class PackData(object):
64
65 def __init__(self, features, labels, batch_size=None, random_shuffle=True):
66 self.__features = features
67 self.__labels = labels
68 self.__batch_size = batch_size
69 self.__random_shuffle = random_shuffle
70
71 self.num = self.__features.shape[0]
72 if self.__batch_size is None:
73 self.__batch_size = self.num
74
75 self.__indices = list(range(self.num))
76 if self.__random_shuffle:
77 numpy.random.shuffle(self.__indices)
78
79 def __call__(self):
80 for i in range(0, self.num, self.__batch_size):
81 batchIndices = self.__indices[i:min(i+self.__batch_size, self.num)]
82 yield self.__features[batchIndices], self.__labels[batchIndices]
83
84
85 # 3. 构建模型: multi-layer perceptron
86 class MLP(object):
87
88 def __init__(self, hidden_dim=100):
89 self.__hidden_dim = hidden_dim
90
91 self.l1_W = torch.normal(0, 0.01, (3, self.__hidden_dim), requires_grad=True)
92 self.l1_b = torch.zeros((1, self.__hidden_dim), requires_grad=True)
93 self.l1_f = torch.nn.Tanh()
94
95 self.l2_W = torch.normal(0, 0.01, (self.__hidden_dim, 3), requires_grad=True)
96 self.l2_b = torch.zeros((1, 3), requires_grad=True)
97
98 def __call__(self, x):
99 l1_1 = torch.matmul(x, self.l1_W) + self.l1_b
100 l1_2 = self.l1_f(l1_1)
101
102 l2_1 = torch.matmul(l1_2, self.l2_W) + self.l2_b
103 return l2_1
104
105
106 # 4. 构建损失函数
107 class MSE(object):
108
109 def __init__(self, lamda):
110 self.__lamda = lamda
111
112 def __call__(self, Y, Y_, mlpObj=None):
113 L = torch.sum((Y - Y_) ** 2) / 2
114 if mlpObj:
115 term1 = torch.sum(mlpObj.l1_W ** 2)
116 term2 = torch.sum(mlpObj.l2_W ** 2)
117 term3 = (term1 + term2) * self.__lamda / 2
118 L = L + term3
119 return L
120
121
122 # 6. 训练单元
123 def training_epoch(packObj, mlpObj, mseObj, optObj):
124 loss_total = 0
125 with torch.enable_grad():
126 for X, Y_ in packObj():
127 optObj.zero_grad()
128 Y = mlpObj(X)
129 loss = mseObj(Y, Y_, mlpObj)
130 loss.backward()
131 optObj.step()
132
133 loss_total += loss.item()
134 return loss_total
135
136
137 # 7. 测试单元
138 def testing_epoch(packObj, mlpObj, mseObj):
139 loss_total = 0
140 with torch.no_grad():
141 for X, Y_ in packObj():
142 Y = mlpObj(X)
143 loss = mseObj(Y, Y_)
144 loss_total += loss.item()
145 return loss_total
146
147
148 # 8. 启动训练与测试
149 def train(trainingData, testingData, model, loss, optimizer, maxEpoch=10000):
150 testingLossList = list()
151 for epoch in range(maxEpoch):
152 training_epoch(trainingData, model, loss, optimizer)
153 testingLoss = testing_epoch(testingData, model, loss) / testingData.num
154 testingLossList.append(testingLoss)
155 # if epoch % 100 == 0:
156 # print("epoch {}: testing error = {:.5f}".format(epoch,
157 # testingLoss))
158
159 minIdx = numpy.argmin(testingLossList)
160 testingLossBest = testingLossList[minIdx]
161 return testingLossBest
162
163
164 # 9. 模型保存
165 def save(model, filename=None):
166 l1_W = model.l1_W.detach().numpy()
167 l1_b = model.l1_b.detach().numpy()
168 l2_W = model.l2_W.detach().numpy()
169 l2_b = model.l2_b.detach().numpy()
170
171 if filename is None:
172 filename = "./mlp.dat"
173 with open(filename, "wt") as f:
174 f.write("l1_W = \n")
175 for row in l1_W:
176 for ele in row:
177 f.write("{:.9f} ".format(ele))
178 f.write("\n")
179 f.write("\nl1_b = \n")
180 for ele in l1_b[0]:
181 f.write("{:.9f} ".format(ele))
182 f.write("\n")
183
184 f.write("\nl2_W = \n")
185 for row in l2_W:
186 for ele in row:
187 f.write("{:.9f} ".format(ele))
188 f.write("\n")
189 f.write("\nl2_b = \n")
190 for ele in l2_b[0]:
191 f.write("{:.9f} ".format(ele))
192
193
194 # 搜索超参数lamda
195 def search_lamda():
196 rRange = [-10, 10]
197 gRange = [-10, 10]
198 bRange = [-10, 10]
199 trainingNum = 500
200 testingNum = 1000
201 batch_size = 250
202 hidden_dim = 50
203
204 geneObj = GeneData(rRange, gRange, bRange)
205 trainingData = geneObj.getDataset(trainingNum)
206 testingData = geneObj.getDataset(testingNum)
207 trainingPack = PackData(*trainingData, batch_size)
208 testingPack = PackData(*testingData, batch_size)
209
210 lamda = 0.001
211 lr = 0.003
212 mlpObj = MLP(hidden_dim)
213 mseObj = MSE(lamda)
214 params = [mlpObj.l1_W, mlpObj.l1_b, mlpObj.l2_W, mlpObj.l2_b]
215 optObj = optim.Adam(params, lr)
216 train(trainingPack, testingPack, mlpObj, mseObj, optObj, 100000)
217 l1_W, l1_b, l2_W, l2_b = mlpObj.l1_W, mlpObj.l1_b, mlpObj.l2_W, mlpObj.l2_b
218
219 lr = 0.003
220 lamdaList = numpy.linspace(0, 0.01, 101)
221 testList = list()
222 for idx, lamda in enumerate(lamdaList):
223 mlpObj = MLP(hidden_dim)
224 mlpObj.l1_W.requires_grad = False
225 mlpObj.l1_b.requires_grad = False
226 mlpObj.l2_W.requires_grad = False
227 mlpObj.l2_b.requires_grad = False
228 l1_W.requires_grad = False
229 l1_b.requires_grad = False
230 l2_W.requires_grad = False
231 l2_b.requires_grad = False
232 mlpObj.l1_W[:], mlpObj.l1_b[:], mlpObj.l2_W[:], mlpObj.l2_b[:] = l1_W, l1_b, l2_W, l2_b
233 mlpObj.l1_W.requires_grad = True
234 mlpObj.l1_b.requires_grad = True
235 mlpObj.l2_W.requires_grad = True
236 mlpObj.l2_b.requires_grad = True
237 mseObj = MSE(lamda)
238 params = [mlpObj.l1_W, mlpObj.l1_b, mlpObj.l2_W, mlpObj.l2_b]
239 optObj = optim.Adam(params, lr)
240 testingLoss = train(trainingPack, testingPack, mlpObj, mseObj, optObj, 100000)
241 print("lamda = {:5f}, testing error = {}".format(lamda, testingLoss))
242 testList.append(testingLoss)
243 l1_W, l1_b, l2_W, l2_b = mlpObj.l1_W, mlpObj.l1_b, mlpObj.l2_W, mlpObj.l2_b
244
245 minIdx = numpy.argmin(testList)
246 lamdaBest = lamdaList[minIdx]
247 testBest = testList[minIdx]
248
249 fig = plt.figure(figsize=(5, 4))
250 ax1 = fig.add_subplot(1, 1, 1)
251 ax1.plot(lamdaList, testList, ".--", lw=1, markersize=5, label="testing error", zorder=1)
252 ax1.scatter(lamdaBest, testBest, marker="*", s=30, c="red", label="optimal", zorder=2)
253 ax1.set(xlabel="$\\lambda$", ylabel="error", title="optimal $\\lambda$ = {:.5f}".format(lamdaBest))
254 ax1.legend()
255 fig.tight_layout()
256 fig.savefig("search_lamda.png", dpi=100)
257
258 ############
259 maxEpoch = 100000
260 mlpObj = MLP(hidden_dim)
261 mseObj = MSE(lamdaBest)
262 params = [mlpObj.l1_W, mlpObj.l1_b, mlpObj.l2_W, mlpObj.l2_b]
263 optObj = optim.Adam(params, lr)
264
265 testingLossBest = numpy.inf
266 for epoch in range(maxEpoch):
267 training_epoch(trainingPack, mlpObj, mseObj, optObj)
268 testingLoss = testing_epoch(testingPack, mlpObj, mseObj) / testingPack.num
269 print("epoch {}: testing error best = {}, testing error current = {}".format(epoch, testingLossBest, testingLoss))
270 if testingLoss < testingLossBest:
271 save(mlpObj)
272 testingLossBest = testingLoss
273
274
275
276 if __name__ == "__main__":
277 search_lamda()
 View Code
View Code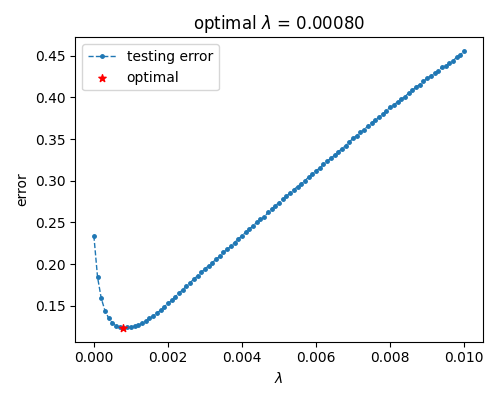 可以看到, 泛化误差在提升权重参数后先下降后上升, 大致对应降低模型复杂度使模型表现从过拟合至欠拟合.
可以看到, 泛化误差在提升权重参数后先下降后上升, 大致对应降低模型复杂度使模型表现从过拟合至欠拟合.