0.先说一下本机配置
opencv2+cuda7.5+cudnn+anaconda,这些基础的之前都是配置好了的,python环境建议使用anaconda,用到的库基本都有了,好像没有easydict,自己装一下就好。
1.下载py-faster-rcnn
rbg大神github上的py-faster-rcnn
使用以下命令下载,直接download的话不完整
git clone --recursive https://github.com/rbgishick/py-faster-rcnn.git
2.编译lib
因为原版是在linux中使用的,在linux里可以直接编译,在windows下需要修改lib/setup.py,以下是修改后的
# --------------------------------------------------------
# Fast R-CNN
# Copyright (c) 2015 Microsoft
# Licensed under The MIT License [see LICENSE for details]
# Written by Ross Girshick
# --------------------------------------------------------
import numpy as np
import os
from os.path import join as pjoin
#from distutils.core import setup
from setuptools import setup
from distutils.extension import Extension
from Cython.Distutils import build_ext
import subprocess
#change for windows, by MrX
nvcc_bin = 'nvcc.exe'
lib_dir = 'lib/x64'
def find_in_path(name, path):
"Find a file in a search path"
# Adapted fom
# http://code.activestate.com/recipes/52224-find-a-file-given-a-search-path/
for dir in path.split(os.pathsep):
binpath = pjoin(dir, name)
if os.path.exists(binpath):
return os.path.abspath(binpath)
return None
def locate_cuda():
"""Locate the CUDA environment on the system
Returns a dict with keys 'home', 'nvcc', 'include', and 'lib64'
and values giving the absolute path to each directory.
Starts by looking for the CUDAHOME env variable. If not found, everything
is based on finding 'nvcc' in the PATH.
"""
# first check if the CUDAHOME env variable is in use
if 'CUDA_PATH' in os.environ:
home = os.environ['CUDA_PATH']
print("home = %s
" % home)
nvcc = pjoin(home, 'bin', nvcc_bin)
else:
# otherwise, search the PATH for NVCC
default_path = pjoin(os.sep, 'usr', 'local', 'cuda', 'bin')
nvcc = find_in_path(nvcc_bin, os.environ['PATH'] + os.pathsep + default_path)
if nvcc is None:
raise EnvironmentError('The nvcc binary could not be '
'located in your $PATH. Either add it to your path, or set $CUDA_PATH')
home = os.path.dirname(os.path.dirname(nvcc))
print("home = %s, nvcc = %s
" % (home, nvcc))
cudaconfig = {'home':home, 'nvcc':nvcc,
'include': pjoin(home, 'include'),
'lib64': pjoin(home, lib_dir)}
for k, v in cudaconfig.iteritems():
if not os.path.exists(v):
raise EnvironmentError('The CUDA %s path could not be located in %s' % (k, v))
return cudaconfig
CUDA = locate_cuda()
# Obtain the numpy include directory. This logic works across numpy versions.
try:
numpy_include = np.get_include()
except AttributeError:
numpy_include = np.get_numpy_include()
def customize_compiler_for_nvcc(self):
"""inject deep into distutils to customize how the dispatch
to cl/nvcc works.
If you subclass UnixCCompiler, it's not trivial to get your subclass
injected in, and still have the right customizations (i.e.
distutils.sysconfig.customize_compiler) run on it. So instead of going
the OO route, I have this. Note, it's kindof like a wierd functional
subclassing going on."""
# tell the compiler it can processes .cu
#self.src_extensions.append('.cu')
# save references to the default compiler_so and _comple methods
#default_compiler_so = self.spawn
#default_compiler_so = self.rc
super = self.compile
# now redefine the _compile method. This gets executed for each
# object but distutils doesn't have the ability to change compilers
# based on source extension: we add it.
def compile(sources, output_dir=None, macros=None, include_dirs=None, debug=0, extra_preargs=None, extra_postargs=None, depends=None):
postfix=os.path.splitext(sources[0])[1]
if postfix == '.cu':
# use the cuda for .cu files
#self.set_executable('compiler_so', CUDA['nvcc'])
# use only a subset of the extra_postargs, which are 1-1 translated
# from the extra_compile_args in the Extension class
postargs = extra_postargs['nvcc']
else:
postargs = extra_postargs['cl']
return super(sources, output_dir, macros, include_dirs, debug, extra_preargs, postargs, depends)
# reset the default compiler_so, which we might have changed for cuda
#self.rc = default_compiler_so
# inject our redefined _compile method into the class
self.compile = compile
# run the customize_compiler
class custom_build_ext(build_ext):
def build_extensions(self):
customize_compiler_for_nvcc(self.compiler)
build_ext.build_extensions(self)
ext_modules = [
# unix _compile: obj, src, ext, cc_args, extra_postargs, pp_opts
Extension(
"utils.cython_bbox",
sources=["utils\bbox.pyx"],
#define_macros={'/LD'},
#extra_compile_args={'cl': ['/link', '/DLL', '/OUT:cython_bbox.dll']},
#extra_compile_args={'cl': ['/LD']},
extra_compile_args={'cl': []},
include_dirs = [numpy_include]
),
Extension(
"nms.cpu_nms",
sources=["nms\cpu_nms.pyx"],
extra_compile_args={'cl': []},
include_dirs = [numpy_include],
),
Extension(
'pycocotools._mask',
sources=['pycocotools\maskApi.c', 'pycocotools\_mask.pyx'],
include_dirs = [numpy_include, 'pycocotools'],
extra_compile_args={'cl': []},
),
#Extension( # just used to get nmsgpu_nms.obj
# "nms.gpu_nms",
# sources=['nms\gpu_nms.pyx'],
# language='c++',
# extra_compile_args={'cl': []},
# include_dirs = [numpy_include]
#),
]
setup(
name='fast_rcnn',
ext_modules=ext_modules,
# inject our custom trigger
cmdclass={'build_ext': custom_build_ext},
)
在lib文件夹中添加setup_cuda.py,内容如下:
#!/usr/bin/env python
import numpy as np
import os
# on Windows, we need the original PATH without Anaconda's compiler in it:
PATH = os.environ.get('PATH')
from distutils.spawn import spawn, find_executable
from setuptools import setup, find_packages, Extension
from setuptools.command.build_ext import build_ext
import sys
# CUDA specific config
# nvcc is assumed to be in user's PATH
nvcc_compile_args = ['-O', '--ptxas-options=-v', '-arch=sm_35', '-c', '--compiler-options=-fPIC']
nvcc_compile_args = os.environ.get('NVCCFLAGS', '').split() + nvcc_compile_args
cuda_libs = ['cublas']
# Obtain the numpy include directory. This logic works across numpy versions.
try:
numpy_include = np.get_include()
except AttributeError:
numpy_include = np.get_numpy_include()
cudamat_ext = Extension('nms.gpu_nms',
sources=[
'nms\gpu_nms.cu'
],
language='c++',
libraries=cuda_libs,
extra_compile_args=nvcc_compile_args,
include_dirs = [numpy_include, 'C:\Programming\CUDA\v7.5\include'])
class CUDA_build_ext(build_ext):
"""
Custom build_ext command that compiles CUDA files.
Note that all extension source files will be processed with this compiler.
"""
def build_extensions(self):
self.compiler.src_extensions.append('.cu')
self.compiler.set_executable('compiler_so', 'nvcc')
self.compiler.set_executable('linker_so', 'nvcc --shared')
if hasattr(self.compiler, '_c_extensions'):
self.compiler._c_extensions.append('.cu') # needed for Windows
self.compiler.spawn = self.spawn
build_ext.build_extensions(self)
def spawn(self, cmd, search_path=1, verbose=0, dry_run=0):
"""
Perform any CUDA specific customizations before actually launching
compile/link etc. commands.
"""
if (sys.platform == 'darwin' and len(cmd) >= 2 and cmd[0] == 'nvcc' and
cmd[1] == '--shared' and cmd.count('-arch') > 0):
# Versions of distutils on OSX earlier than 2.7.9 inject
# '-arch x86_64' which we need to strip while using nvcc for
# linking
while True:
try:
index = cmd.index('-arch')
del cmd[index:index+2]
except ValueError:
break
elif self.compiler.compiler_type == 'msvc':
# There are several things we need to do to change the commands
# issued by MSVCCompiler into one that works with nvcc. In the end,
# it might have been easier to write our own CCompiler class for
# nvcc, as we're only interested in creating a shared library to
# load with ctypes, not in creating an importable Python extension.
# - First, we replace the cl.exe or link.exe call with an nvcc
# call. In case we're running Anaconda, we search cl.exe in the
# original search path we captured further above -- Anaconda
# inserts a MSVC version into PATH that is too old for nvcc.
cmd[:1] = ['nvcc', '--compiler-bindir',
os.path.dirname(find_executable("cl.exe", PATH))
or cmd[0]]
# - Secondly, we fix a bunch of command line arguments.
for idx, c in enumerate(cmd):
# create .dll instead of .pyd files
#if '.pyd' in c: cmd[idx] = c = c.replace('.pyd', '.dll') #20160601, by MrX
# replace /c by -c
if c == '/c': cmd[idx] = '-c'
# replace /DLL by --shared
elif c == '/DLL': cmd[idx] = '--shared'
# remove --compiler-options=-fPIC
elif '-fPIC' in c: del cmd[idx]
# replace /Tc... by ...
elif c.startswith('/Tc'): cmd[idx] = c[3:]
# replace /Fo... by -o ...
elif c.startswith('/Fo'): cmd[idx:idx+1] = ['-o', c[3:]]
# replace /LIBPATH:... by -L...
elif c.startswith('/LIBPATH:'): cmd[idx] = '-L' + c[9:]
# replace /OUT:... by -o ...
elif c.startswith('/OUT:'): cmd[idx:idx+1] = ['-o', c[5:]]
# remove /EXPORT:initlibcudamat or /EXPORT:initlibcudalearn
elif c.startswith('/EXPORT:'): del cmd[idx]
# replace cublas.lib by -lcublas
elif c == 'cublas.lib': cmd[idx] = '-lcublas'
# - Finally, we pass on all arguments starting with a '/' to the
# compiler or linker, and have nvcc handle all other arguments
if '--shared' in cmd:
pass_on = '--linker-options='
# we only need MSVCRT for a .dll, remove CMT if it sneaks in:
cmd.append('/NODEFAULTLIB:libcmt.lib')
else:
pass_on = '--compiler-options='
cmd = ([c for c in cmd if c[0] != '/'] +
[pass_on + ','.join(c for c in cmd if c[0] == '/')])
# For the future: Apart from the wrongly set PATH by Anaconda, it
# would suffice to run the following for compilation on Windows:
# nvcc -c -O -o <file>.obj <file>.cu
# And the following for linking:
# nvcc --shared -o <file>.dll <file1>.obj <file2>.obj -lcublas
# This could be done by a NVCCCompiler class for all platforms.
spawn(cmd, search_path, verbose, dry_run)
setup(name="py_fast_rcnn_gpu",
description="Performs linear algebra computation on the GPU via CUDA",
ext_modules=[cudamat_ext],
cmdclass={'build_ext': CUDA_build_ext},
)
修改之后就可以编译了,将CMD定位到py-faster-rcnn目录下的lib文件夹,运行一下命令:
python setup.py install
python setup_cuda.py install
如果提示Microsoft Visual C++ 9.0 is required ...,在CMD中输入以下命令:
SET VS90COMNTOOLS=%VS110COMNTOOLS% (如果电脑中装的是vs2012)
SET VS90COMNTOOLS=%VS120COMNTOOLS% (如果电脑中装的是vs2013)
然后最后出现Finished processing ...就编译成功了。
3.编译pycaffe并替换到py-faster-rcnn目录下
之前已经安装了caffe的windows版,然后要编译pycaffe,打开buildVS2013下的工程,在pycaffe的属性中的预编译器中添加 WITH_PYTHON_LAYER,这样才会有Python类型的layer可以用
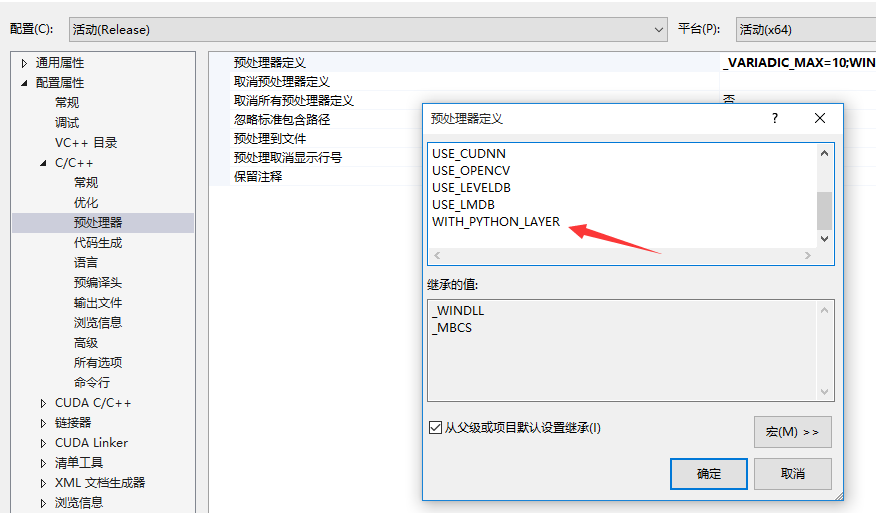
编译后将生成的文件,也就是caffe目录下python/caffe这个文件夹,整体复制到py-faster-rcnn目录下的caffe-fast-rcnn中python文件夹中并替换。
4.下载model文件
在data/scripts/fetch_faster_rcnn_models.sh中有个链接,可以在这里下载可用的models,文件600多兆,下完之后解压到data文件夹下(因为是.tar.gz压缩,记得解压两次),将解压完的文件夹
faster_rcnn_models文件夹放在data目录下
另外,如果想替换成自己的caffemodel,需要修改faster_rcnn_test.pt文件,(在modelspascal_vocVGG16目录下)
5.运行demo.py
然后就可以运行tools下的demo.py了,将CMD定位到tools目录
python demo.py
因为会导入cv2库,所以之前需要将opencv2/build/python/2.7/x64(也可能是x86)下的cv2.pyd复制到当前python的库搜索路径,可以使用sys.path来查看当前的搜索路径。
然后在运行demo.py出现有param_str_这个错误的话,就将lib文件夹下的rpn和roid_data_layer中的所有.py文件中的param_str_改成param_str,因为新版本的caffe中没有最后的下划线。