Classification of text documents using sparse features
https://scikit-learn.org/stable/auto_examples/text/plot_document_classification_20newsgroups.html#sphx-glr-auto-examples-text-plot-document-classification-20newsgroups-py
使用词袋方法进行文档按主题分类,数据对象为20主题新闻数据集。
使用稀疏矩阵存储稀疏特征,使用不同的分类器测试特性。
This is an example showing how scikit-learn can be used to classify documents by topics using a bag-of-words approach.
This example uses a scipy.sparse matrix to store the features and demonstrates various classifiers that can efficiently handle sparse matrices.
The dataset used in this example is the 20 newsgroups dataset. It will be automatically downloaded, then cached.
Code
""" ====================================================== Classification of text documents using sparse features ====================================================== This is an example showing how scikit-learn can be used to classify documents by topics using a bag-of-words approach. This example uses a scipy.sparse matrix to store the features and demonstrates various classifiers that can efficiently handle sparse matrices. The dataset used in this example is the 20 newsgroups dataset. It will be automatically downloaded, then cached. """ # Author: Peter Prettenhofer <peter.prettenhofer@gmail.com> # Olivier Grisel <olivier.grisel@ensta.org> # Mathieu Blondel <mathieu@mblondel.org> # Lars Buitinck # License: BSD 3 clause import logging import numpy as np from optparse import OptionParser import sys from time import time import matplotlib.pyplot as plt from sklearn.datasets import fetch_20newsgroups from sklearn.feature_extraction.text import TfidfVectorizer from sklearn.feature_extraction.text import HashingVectorizer from sklearn.feature_selection import SelectFromModel from sklearn.feature_selection import SelectKBest, chi2 from sklearn.linear_model import RidgeClassifier from sklearn.pipeline import Pipeline from sklearn.svm import LinearSVC from sklearn.linear_model import SGDClassifier from sklearn.linear_model import Perceptron from sklearn.linear_model import PassiveAggressiveClassifier from sklearn.naive_bayes import BernoulliNB, ComplementNB, MultinomialNB from sklearn.neighbors import KNeighborsClassifier from sklearn.neighbors import NearestCentroid from sklearn.ensemble import RandomForestClassifier from sklearn.utils.extmath import density from sklearn import metrics # Display progress logs on stdout logging.basicConfig(level=logging.INFO, format='%(asctime)s %(levelname)s %(message)s') op = OptionParser() op.add_option("--report", action="store_true", dest="print_report", help="Print a detailed classification report.") op.add_option("--chi2_select", action="store", type="int", dest="select_chi2", help="Select some number of features using a chi-squared test") op.add_option("--confusion_matrix", action="store_true", dest="print_cm", help="Print the confusion matrix.") op.add_option("--top10", action="store_true", dest="print_top10", help="Print ten most discriminative terms per class" " for every classifier.") op.add_option("--all_categories", action="store_true", dest="all_categories", help="Whether to use all categories or not.") op.add_option("--use_hashing", action="store_true", help="Use a hashing vectorizer.") op.add_option("--n_features", action="store", type=int, default=2 ** 16, help="n_features when using the hashing vectorizer.") op.add_option("--filtered", action="store_true", help="Remove newsgroup information that is easily overfit: " "headers, signatures, and quoting.") def is_interactive(): return not hasattr(sys.modules['__main__'], '__file__') # work-around for Jupyter notebook and IPython console argv = [] if is_interactive() else sys.argv[1:] (opts, args) = op.parse_args(argv) if len(args) > 0: op.error("this script takes no arguments.") sys.exit(1) print(__doc__) op.print_help() print() # %% # Load data from the training set # ------------------------------------ # Let's load data from the newsgroups dataset which comprises around 18000 # newsgroups posts on 20 topics split in two subsets: one for training (or # development) and the other one for testing (or for performance evaluation). if opts.all_categories: categories = None else: categories = [ 'alt.atheism', 'talk.religion.misc', 'comp.graphics', 'sci.space', ] if opts.filtered: remove = ('headers', 'footers', 'quotes') else: remove = () print("Loading 20 newsgroups dataset for categories:") print(categories if categories else "all") data_train = fetch_20newsgroups(subset='train', categories=categories, shuffle=True, random_state=42, remove=remove) data_test = fetch_20newsgroups(subset='test', categories=categories, shuffle=True, random_state=42, remove=remove) print('data loaded') # order of labels in `target_names` can be different from `categories` target_names = data_train.target_names def size_mb(docs): return sum(len(s.encode('utf-8')) for s in docs) / 1e6 data_train_size_mb = size_mb(data_train.data) data_test_size_mb = size_mb(data_test.data) print("%d documents - %0.3fMB (training set)" % ( len(data_train.data), data_train_size_mb)) print("%d documents - %0.3fMB (test set)" % ( len(data_test.data), data_test_size_mb)) print("%d categories" % len(target_names)) print() # split a training set and a test set y_train, y_test = data_train.target, data_test.target print("Extracting features from the training data using a sparse vectorizer") t0 = time() if opts.use_hashing: vectorizer = HashingVectorizer(stop_words='english', alternate_sign=False, n_features=opts.n_features) X_train = vectorizer.transform(data_train.data) else: vectorizer = TfidfVectorizer(sublinear_tf=True, max_df=0.5, stop_words='english') X_train = vectorizer.fit_transform(data_train.data) duration = time() - t0 print("done in %fs at %0.3fMB/s" % (duration, data_train_size_mb / duration)) print("n_samples: %d, n_features: %d" % X_train.shape) print() print("Extracting features from the test data using the same vectorizer") t0 = time() X_test = vectorizer.transform(data_test.data) duration = time() - t0 print("done in %fs at %0.3fMB/s" % (duration, data_test_size_mb / duration)) print("n_samples: %d, n_features: %d" % X_test.shape) print() # mapping from integer feature name to original token string if opts.use_hashing: feature_names = None else: feature_names = vectorizer.get_feature_names() if opts.select_chi2: print("Extracting %d best features by a chi-squared test" % opts.select_chi2) t0 = time() ch2 = SelectKBest(chi2, k=opts.select_chi2) X_train = ch2.fit_transform(X_train, y_train) X_test = ch2.transform(X_test) if feature_names: # keep selected feature names feature_names = [feature_names[i] for i in ch2.get_support(indices=True)] print("done in %fs" % (time() - t0)) print() if feature_names: feature_names = np.asarray(feature_names) def trim(s): """Trim string to fit on terminal (assuming 80-column display)""" return s if len(s) <= 80 else s[:77] + "..." # %% # Benchmark classifiers # ------------------------------------ # We train and test the datasets with 15 different classification models # and get performance results for each model. def benchmark(clf): print('_' * 80) print("Training: ") print(clf) t0 = time() clf.fit(X_train, y_train) train_time = time() - t0 print("train time: %0.3fs" % train_time) t0 = time() pred = clf.predict(X_test) test_time = time() - t0 print("test time: %0.3fs" % test_time) score = metrics.accuracy_score(y_test, pred) print("accuracy: %0.3f" % score) if hasattr(clf, 'coef_'): print("dimensionality: %d" % clf.coef_.shape[1]) print("density: %f" % density(clf.coef_)) if opts.print_top10 and feature_names is not None: print("top 10 keywords per class:") for i, label in enumerate(target_names): top10 = np.argsort(clf.coef_[i])[-10:] print(trim("%s: %s" % (label, " ".join(feature_names[top10])))) print() if opts.print_report: print("classification report:") print(metrics.classification_report(y_test, pred, target_names=target_names)) if opts.print_cm: print("confusion matrix:") print(metrics.confusion_matrix(y_test, pred)) print() clf_descr = str(clf).split('(')[0] return clf_descr, score, train_time, test_time results = [] for clf, name in ( (RidgeClassifier(tol=1e-2, solver="sag"), "Ridge Classifier"), (Perceptron(max_iter=50), "Perceptron"), (PassiveAggressiveClassifier(max_iter=50), "Passive-Aggressive"), (KNeighborsClassifier(n_neighbors=10), "kNN"), (RandomForestClassifier(), "Random forest")): print('=' * 80) print(name) results.append(benchmark(clf)) for penalty in ["l2", "l1"]: print('=' * 80) print("%s penalty" % penalty.upper()) # Train Liblinear model results.append(benchmark(LinearSVC(penalty=penalty, dual=False, tol=1e-3))) # Train SGD model results.append(benchmark(SGDClassifier(alpha=.0001, max_iter=50, penalty=penalty))) # Train SGD with Elastic Net penalty print('=' * 80) print("Elastic-Net penalty") results.append(benchmark(SGDClassifier(alpha=.0001, max_iter=50, penalty="elasticnet"))) # Train NearestCentroid without threshold print('=' * 80) print("NearestCentroid (aka Rocchio classifier)") results.append(benchmark(NearestCentroid())) # Train sparse Naive Bayes classifiers print('=' * 80) print("Naive Bayes") results.append(benchmark(MultinomialNB(alpha=.01))) results.append(benchmark(BernoulliNB(alpha=.01))) results.append(benchmark(ComplementNB(alpha=.1))) print('=' * 80) print("LinearSVC with L1-based feature selection") # The smaller C, the stronger the regularization. # The more regularization, the more sparsity. results.append(benchmark(Pipeline([ ('feature_selection', SelectFromModel(LinearSVC(penalty="l1", dual=False, tol=1e-3))), ('classification', LinearSVC(penalty="l2"))]))) # %% # Add plots # ------------------------------------ # The bar plot indicates the accuracy, training time (normalized) and test time # (normalized) of each classifier. indices = np.arange(len(results)) results = [[x[i] for x in results] for i in range(4)] clf_names, score, training_time, test_time = results training_time = np.array(training_time) / np.max(training_time) test_time = np.array(test_time) / np.max(test_time) plt.figure(figsize=(12, 8)) plt.title("Score") plt.barh(indices, score, .2, label="score", color='navy') plt.barh(indices + .3, training_time, .2, label="training time", color='c') plt.barh(indices + .6, test_time, .2, label="test time", color='darkorange') plt.yticks(()) plt.legend(loc='best') plt.subplots_adjust(left=.25) plt.subplots_adjust(top=.95) plt.subplots_adjust(bottom=.05) for i, c in zip(indices, clf_names): plt.text(-.3, i, c) plt.show()
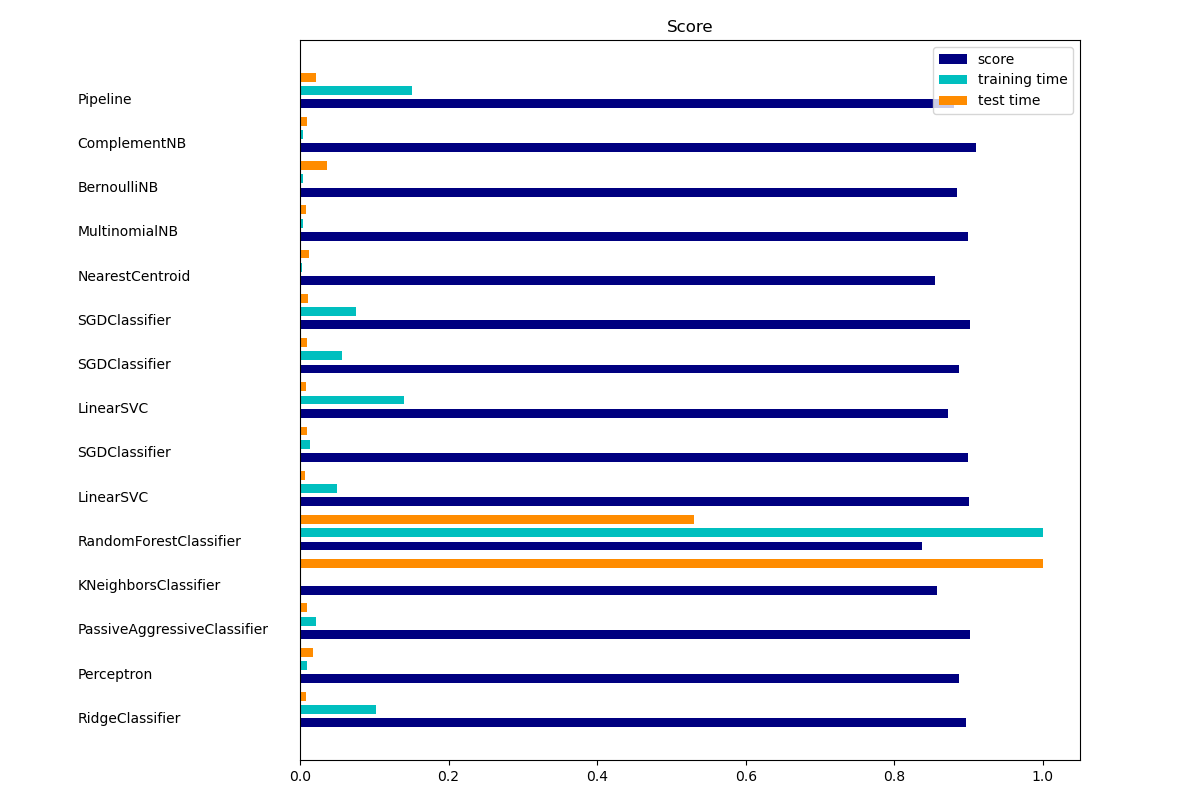
Output
================================================================================
Ridge Classifier
________________________________________________________________________________
Training:
RidgeClassifier(solver='sag', tol=0.01)
/home/circleci/project/sklearn/linear_model/_ridge.py:555: UserWarning: "sag" solver requires many iterations to fit an intercept with sparse inputs. Either set the solver to "auto" or "sparse_cg", or set a low "tol" and a high "max_iter" (especially if inputs are not standardized).
warnings.warn(
train time: 0.180s
test time: 0.001s
accuracy: 0.897
dimensionality: 33809
density: 1.000000
================================================================================
Perceptron
________________________________________________________________________________
Training:
Perceptron(max_iter=50)
train time: 0.017s
test time: 0.003s
accuracy: 0.888
dimensionality: 33809
density: 0.255302
================================================================================
Passive-Aggressive
________________________________________________________________________________
Training:
PassiveAggressiveClassifier(max_iter=50)
train time: 0.039s
test time: 0.002s
accuracy: 0.902
dimensionality: 33809
density: 0.692841
================================================================================
kNN
________________________________________________________________________________
Training:
KNeighborsClassifier(n_neighbors=10)
train time: 0.001s
test time: 0.171s
accuracy: 0.858
================================================================================
Random forest
________________________________________________________________________________
Training:
RandomForestClassifier()
train time: 1.765s
test time: 0.091s
accuracy: 0.837
================================================================================
L2 penalty
________________________________________________________________________________
Training:
LinearSVC(dual=False, tol=0.001)
train time: 0.088s
test time: 0.001s
accuracy: 0.900
dimensionality: 33809
density: 1.000000
________________________________________________________________________________
Training:
SGDClassifier(max_iter=50)
train time: 0.025s
test time: 0.002s
accuracy: 0.899
dimensionality: 33809
density: 0.569944
================================================================================
L1 penalty
________________________________________________________________________________
Training:
LinearSVC(dual=False, penalty='l1', tol=0.001)
train time: 0.247s
test time: 0.001s
accuracy: 0.873
dimensionality: 33809
density: 0.005561
________________________________________________________________________________
Training:
SGDClassifier(max_iter=50, penalty='l1')
train time: 0.099s
test time: 0.002s
accuracy: 0.888
dimensionality: 33809
density: 0.022982
================================================================================
Elastic-Net penalty
________________________________________________________________________________
Training:
SGDClassifier(max_iter=50, penalty='elasticnet')
train time: 0.132s
test time: 0.002s
accuracy: 0.902
dimensionality: 33809
density: 0.187502
================================================================================
NearestCentroid (aka Rocchio classifier)
________________________________________________________________________________
Training:
NearestCentroid()
train time: 0.004s
test time: 0.002s
accuracy: 0.855
================================================================================
Naive Bayes
________________________________________________________________________________
Training:
MultinomialNB(alpha=0.01)
train time: 0.008s
test time: 0.001s
accuracy: 0.899
/home/circleci/project/sklearn/utils/deprecation.py:101: FutureWarning: Attribute coef_ was deprecated in version 0.24 and will be removed in 1.1 (renaming of 0.26).
warnings.warn(msg, category=FutureWarning)
dimensionality: 33809
density: 1.000000
________________________________________________________________________________
Training:
BernoulliNB(alpha=0.01)
train time: 0.008s
test time: 0.006s
accuracy: 0.884
/home/circleci/project/sklearn/utils/deprecation.py:101: FutureWarning: Attribute coef_ was deprecated in version 0.24 and will be removed in 1.1 (renaming of 0.26).
warnings.warn(msg, category=FutureWarning)
dimensionality: 33809
density: 1.000000
________________________________________________________________________________
Training:
ComplementNB(alpha=0.1)
train time: 0.008s
test time: 0.002s
accuracy: 0.911
/home/circleci/project/sklearn/utils/deprecation.py:101: FutureWarning: Attribute coef_ was deprecated in version 0.24 and will be removed in 1.1 (renaming of 0.26).
warnings.warn(msg, category=FutureWarning)
dimensionality: 33809
density: 1.000000
================================================================================
LinearSVC with L1-based feature selection
________________________________________________________________________________
Training:
Pipeline(steps=[('feature_selection',
SelectFromModel(estimator=LinearSVC(dual=False, penalty='l1',
tol=0.001))),
('classification', LinearSVC())])
train time: 0.265s
test time: 0.004s
accuracy: 0.880
SelectKBest
https://scikit-learn.org/stable/modules/generated/sklearn.feature_selection.SelectKBest.html
Select features according to the k highest scores.
See also
f_classifANOVA F-value between label/feature for classification tasks.
mutual_info_classifMutual information for a discrete target.
chi2Chi-squared stats of non-negative features for classification tasks.
f_regressionF-value between label/feature for regression tasks.
mutual_info_regressionMutual information for a continuous target.
SelectPercentileSelect features based on percentile of the highest scores.
SelectFprSelect features based on a false positive rate test.
SelectFdrSelect features based on an estimated false discovery rate.
SelectFweSelect features based on family-wise error rate.
GenericUnivariateSelectUnivariate feature selector with configurable mode.
>>> from sklearn.datasets import load_digits >>> from sklearn.feature_selection import SelectKBest, chi2 >>> X, y = load_digits(return_X_y=True) >>> X.shape (1797, 64) >>> X_new = SelectKBest(chi2, k=20).fit_transform(X, y) >>> X_new.shape (1797, 20)
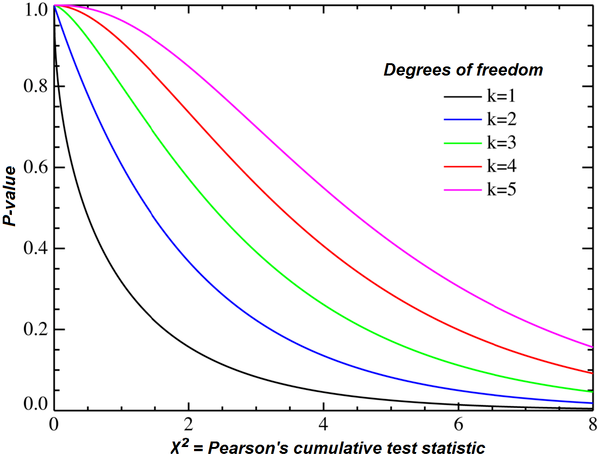
Chi-squared test
对于一个null假设, 假设目标分布跟某一个特征没有显著差异。如果没有显著差异, 则词特征对目标没有更多的贡献,则可以考虑抛弃。
通过卡方检验判定,是否没有显著差异。
卡方检验, 通过计算卡方来查询p值, 重点是计算卡方值, 然后通过公式计算p值, 或者查表法查出p值。
p值越大, 表示null假设越是可信度高,则特征越是没有贡献, p表示possible。
p值越小, 越是需要关注其对目标的贡献度。
https://en.wikipedia.org/wiki/Chi-squared_test
A chi-squared test, also written as χ2 test, is a statistical hypothesis test that is valid to perform when the test statistic is chi-squared distributed under the null hypothesis, specifically Pearson's chi-squared test and variants thereof. Pearson's chi-squared test is used to determine whether there is a statistically significant difference between the expected frequencies and the observed frequencies in one or more categories of a contingency table.
In the standard applications of this test, the observations are classified into mutually exclusive classes. If the null hypothesis that there are no differences between the classes in the population is true, the test statistic computed from the observations follows a χ2 frequency distribution. The purpose of the test is to evaluate how likely the observed frequencies would be assuming the null hypothesis is true.
https://www.mathsisfun.com/data/chi-square-test.html
Example: "Which holiday do you prefer?"
Beach Cruise Men 209 280 Women 225 248 Does Gender affect Preferred Holiday?
If Gender (Man or Woman) does affect Preferred Holiday we say they are dependent.
By doing some special calculations (explained later), we come up with a "p" value:
p value is 0.132
Now, p < 0.05 is the usual test for dependence.
In this case p is greater than 0.05, so we believe the variables are independent (ie not linked together).
In other words Men and Women probably do not have a different preference for Beach Holidays or Cruises.
It was just random differences which we expect when collecting data.
is_interactive
如何判断python是运行在命令行模式, 还是 交互式 运行模式?
只要判断__main__模块中是否包含 __file__ 变量即可。
命令行模式, 代码是从文件中加载进来, 所以有 __file__ 变量。
但是 交互模式, 其运行模式是 REPL , 代码是从用户输入得到, 没有所谓的文件。
def is_interactive():
return not hasattr(sys.modules['__main__'], '__file__')
INTERACTIVE MODE
The interactive mode of Python is also called REPL.
REPL stands for ‘Read-Eval-Print-Loop’. It is a simple, interactive command-line shell that provides us with the result when provided with a single line Python commands.
- Read: The read function accepts an input from the user and stores it into the memory.
- Eval: The eval function evaluates this ‘input’ read from the memory.
- Print: The print function prints the outcome from the eval function.
- Loop: The loop function creates a loop and terminates itself when the program ends.
This was a brief explanation of the interactive mode: REPL.
__file__
https://docs.python.org/3/reference/import.html#__file__
__file__is optional. If set, this attribute’s value must be a string. The import system may opt to leave__file__unset if it has no semantic meaning (e.g. a module loaded from a database).If
__file__is set, it may also be appropriate to set the__cached__attribute which is the path to any compiled version of the code (e.g. byte-compiled file). The file does not need to exist to set this attribute; the path can simply point to where the compiled file would exist (see PEP 3147).It is also appropriate to set
__cached__when__file__is not set. However, that scenario is quite atypical. Ultimately, the loader is what makes use of__file__and/or__cached__. So if a loader can load from a cached module but otherwise does not load from a file, that atypical scenario may be appropriate.
https://stackoverflow.com/questions/28569328/python-main-file
When you run the script from a file then the
__main__module is in fact that file. In the Python interpreter prompt, on the other hand, the__main__module is just the default namespace theinterpreter, specifically the interactive prompt is running in, and it has no file associated with it (loosely speaking the file is<stdin>).When you hit F5 to run the code in idle it's doing the equivalent to if you just copy/pasted that code into the interpreter directly. There is no way for it to have any association with that file.
If, on the other hand, you run
import Test_then now the code in that file is associated with theTest_module and you'll find thatTest_.__file__gives the associated filename.For what it's worth, there's almost never any reason to
import __main__. If you want the script to print the file it was run out of you can just have:if __name__ == '__main__': print __file__among other possibilities.