GO是一个同源蛋白的数据库,按照三大类别BP(生物学过程), MF(分子功能), CC(细胞组分) 对基因的产物-蛋白质进行了分类;
在GO数据库中,本质上是一个有向无环图的数据结构,在三大类别之下,又有小的分类层级,一层一层的分类下去;
对于某个具体的GO号来说,代表了一组同源蛋白,拥有相似的结构和功能,在数据库中,有上层分类的GO,也可能在这个GO下面,还有其他的GO
为了直观的展示某个GO号的分类,利用数据库提供的go-basic.obo 文件得到不同GO之间的层级关系,然后用Graphviz 进行可视化
首先下载go-basic.obo 文件, 到GO官网 http://geneontology.org/page/download-ontology

选择下载go-basic.obo 文件
wget http://purl.obolibrary.org/obo/go/go-basic.obo
观察obo 文件
[Term] id: GO:0000001 name: mitochondrion inheritance namespace: biological_process def: "The distribution of mitochondria, including the mitochondrial genome, into daughter cells after mitosis or meiosis, mediated by interaction synonym: "mitochondrial inheritance" EXACT [] is_a: GO:0048308 ! organelle inheritance is_a: GO:0048311 ! mitochondrion distribution
其核心是Term , 每个Term 下记录了一个具体的GO, name 是对该GO的简单描述,namespace 是值该GO 属于三大类别中的哪一个,def 是详细的描述信息, is_a 记录了该GO 对应的上层分类的GO
根据每个GO的is_a的信息,就可以得到所有GO之间的相互关系,利用程序处理,对于某个具体的GO,就可以得到其对应的所有上层分类,然后整理成Graphviz 识别的dot 文件,进行可视化
具体的代码如下:
#!/usr/bin/env perl my ($obo, $go) = @ARGV; my %nodes = (); my %edges = (); local $/ = "[Term]"; open OBO, $obo or die "Can't open $obo! "; while (<OBO>) { chomp; my ($node) = $_ =~ /id:s+(S+)/; next if not defined $node; my ($name) = $_ =~ /name:s+(.+?) /; my $label = qq{"$node\n$name"}; $node =~ s/://; $nodes{$node} = $label; my @edge = $_ =~ /is_a:s+(GO:d+)/; foreach my $x (@edge) { $x =~ s/://; push @{$edges{$node}}, $x; } } close OBO; my @val = @{$edges{$go}}; my @node = (); my @edge = (); push @node, $go; my $start = $go; while (@val) { my $id = shift @val; push @node, $id; push @edge, qq{$id -> $start;}; foreach my $x (@{$edges{$id}}) { push @val, $x; } $start = $id; } print qq{digraph go{n}; foreach my $x (@node) { print qq{$x[label = $nodes{$x}];n}; } foreach my $x (@edge) { print qq{$xn}; } print qq{}};
运行的命令如下:
perl parse_go.pl go-basic.obo GO0002485 > GO0002485.dot
如果使用GO:0002485 这种作为结点的名字,Graphviz 不能够正确识别,所以我把冒号统一替换掉了,生成的GO0002485的内容如下:
digraph go{
GO0002485[label = "GO:0002485
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent"];
GO0002484[label = "GO:0002484
antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway"];
GO0019885[label = "GO:0019885
antigen processing and presentation of endogenous peptide antigen via MHC class I"];
GO0002474[label = "GO:0002474
antigen processing and presentation of peptide antigen via MHC class I"];
GO0048002[label = "GO:0048002
antigen processing and presentation of peptide antigen"];
GO0019882[label = "GO:0019882
antigen processing and presentation"];
GO0002376[label = "GO:0002376
immune system process"];
GO0008150[label = "GO:0008150
biological_process"];
GO0002484 -> GO0002485;
GO0019885 -> GO0002484;
GO0002474 -> GO0019885;
GO0048002 -> GO0002474;
GO0019882 -> GO0048002;
GO0002376 -> GO0019882;
GO0008150 -> GO0002376;
}
最后用Graphviz 进行可视化
dot -T pdf GO0002485.dot -o GO0002485.pdf
效果图如下:
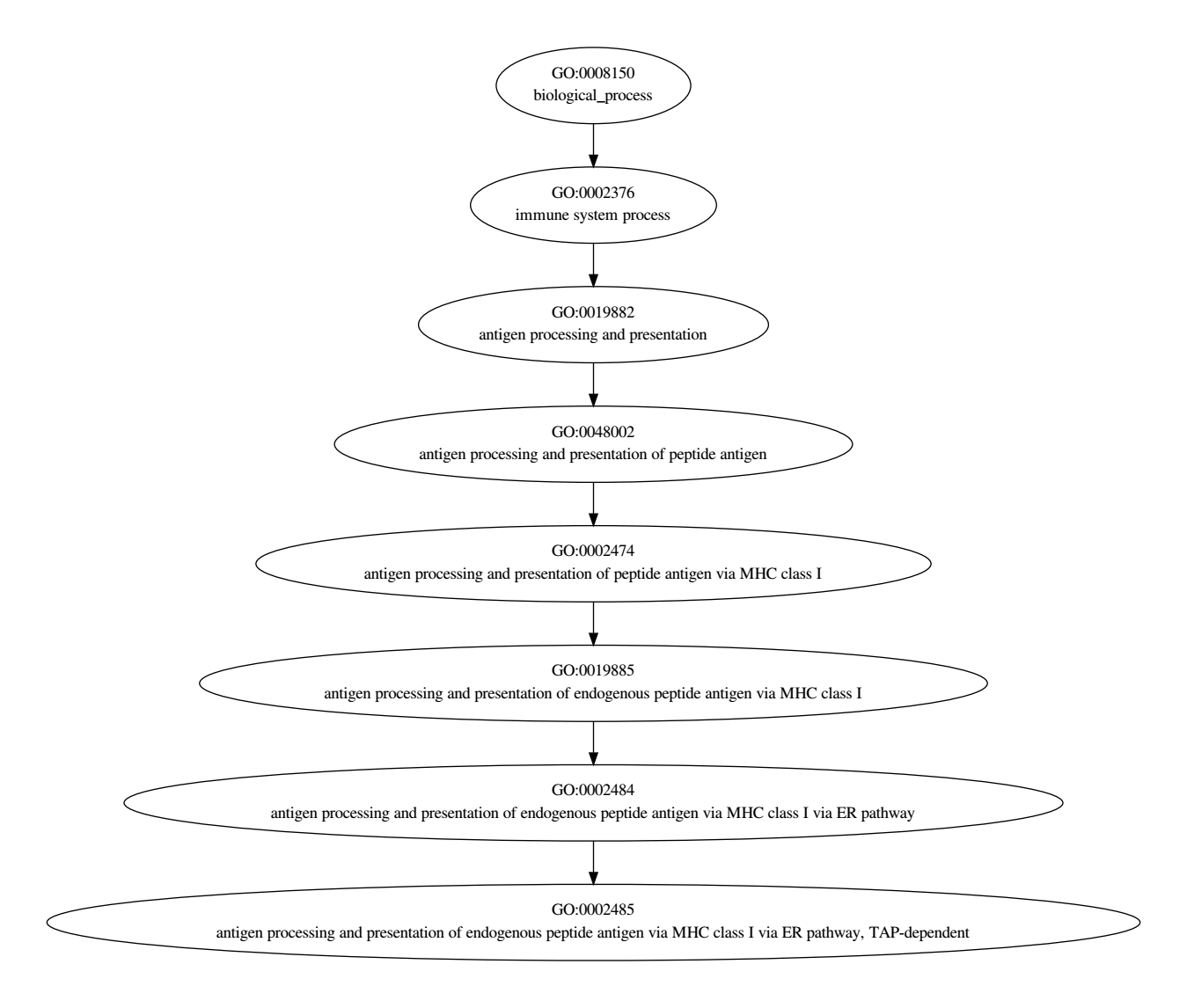
这里全部采用了默认的格式,其实还可以在这个基础上再修改样式,使图片更加的美观;