001、
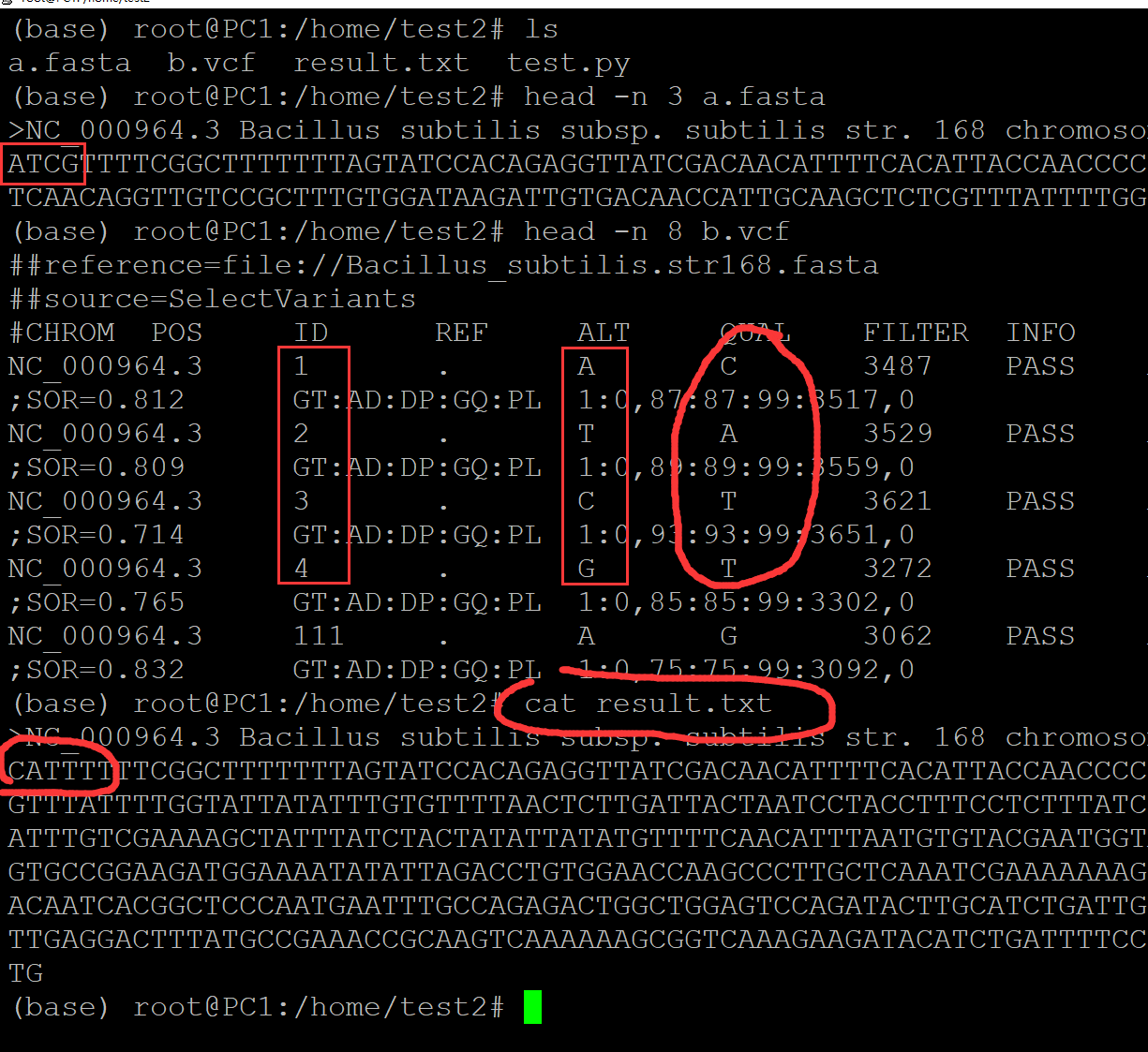
(base) root@PC1:/home/test2# ls a.fasta b.vcf test.py (base) root@PC1:/home/test2# cat a.fasta ## 测试fasta文件 >NC_000964.3 Bacillus subtilis subsp. subtilis str. 168 chromosome, complete genome ATCGTTTTCGGCTTTTTTTAGTATCCACAGAGGTTATCGACAACATTTTCACATTACCAACCCCTGTGGACAAGGTTTTT TCAACAGGTTGTCCGCTTTGTGGATAAGATTGTGACAACCATTGCAAGCTCTCGTTTATTTTGGTATTATATTTGTGTTT TAACTCTTGATTACTAATCCTACCTTTCCTCTTTATCCACAAAGTGTGGATAAGTTGTGGATTGATTTCACACAGCTTGT GTAGAAGGTTGTCCACAAGTTGTGAAATTTGTCGAAAAGCTATTTATCTACTATATTATATGTTTTCAACATTTAATGTG TACGAATGGTAAGCGCCATTTGCTCTTTTTTTGTGTTCTATAACAGAGAAAGACGCCATTTTCTAAGAAAAGGAGGGACG TGCCGGAAGATGGAAAATATATTAGACCTGTGGAACCAAGCCCTTGCTCAAATCGAAAAAAAGTTGAGCAAACCGAGTTT TGAGACTTGGATGAAGTCAACCAAAGCCCACTCACTGCAAGGCGATACATTAACAATCACGGCTCCCAATGAATTTGCCA GAGACTGGCTGGAGTCCAGATACTTGCATCTGATTGCAGATACTATATATGAATTAACCGGGGAAGAATTGAGCATTAAG TTTGTCATTCCTCAAAATCAAGATGTTGAGGACTTTATGCCGAAACCGCAAGTCAAAAAAGCGGTCAAAGAAGATACATC TGATTTTCCTCAAAATATGCTCAATCCAAAATATACTTTTGATACTTTTGTCATCGGATCTGGAAACCGATTTGCACATG (base) root@PC1:/home/test2# cat b.vcf ## 测试vcf文件 ##reference=file://Bacillus_subtilis.str168.fasta ##source=SelectVariants #CHROM POS ID REF ALT QUAL FILTER INFO FORMAT Bacillus_subtilis NC_000964.3 1 . A C 3487 PASS AC=1;AF=1.00;AN=1;DP=87;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=34.24;SOR=0.812 GT:AD:DP:GQ:PL 1:0,87:87:99:3517,0 NC_000964.3 2 . T A 3529 PASS AC=1;AF=1.00;AN=1;DP=89;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=30.63;SOR=0.809 GT:AD:DP:GQ:PL 1:0,89:89:99:3559,0 NC_000964.3 3 . C T 3621 PASS AC=1;AF=1.00;AN=1;DP=93;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=29.09;SOR=0.714 GT:AD:DP:GQ:PL 1:0,93:93:99:3651,0 NC_000964.3 4 . G T 3272 PASS AC=1;AF=1.00;AN=1;DP=86;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=32.93;SOR=0.765 GT:AD:DP:GQ:PL 1:0,85:85:99:3302,0 NC_000964.3 111 . A G 3062 PASS AC=1;AF=1.00;AN=1;DP=75;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=30.83;SOR=0.832 GT:AD:DP:GQ:PL 1:0,75:75:99:3092,0 NC_000964.3 115 . G A 2962 PASS AC=1;AF=1.00;AN=1;DP=72;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=29.67;SOR=0.807 GT:AD:DP:GQ:PL 1:0,72:72:99:2992,0 NC_000964.3 118 . A G 2805 PASS AC=1;AF=1.00;AN=1;DP=71;FS=0.000;MLEAC=1;MLEAF=1.00;MQ=60.00;QD=34.04;SOR=0.779 GT:AD:DP:GQ:PL 1:0,71:71:99:2835,0 (base) root@PC1:/home/test2# cat test.py ## 测试脚本 #!/usr/bin/python in_file1 = open("a.fasta", "r") in_file2 = open("b.vcf", "r") out_file = open("result.txt", "w") seq_all = {} fasta_id_all = [] scaff_id_all = [] for i in in_file1: i = i.strip() if i[0] == ">": fasta_id_all.append(i) seq = i.split()[0].split(">")[1] scaff_id_all.append(seq) seq_all[seq] = [] else: for j in i: seq_all[seq].append(j) for i in in_file2: i = i.strip() if i[0] == "#": continue else: i = i.split("\t") seq_all[i[0]][int(i[1]) - 1] = i[4] for i in scaff_id_all: print(fasta_id_all[scaff_id_all.index(i)], file = out_file) print("".join(seq_all[i]), file = out_file) in_file1.close() in_file2.close() out_file.close() (base) root@PC1:/home/test2# python test.py ## 执行程序 (base) root@PC1:/home/test2# ls a.fasta b.vcf result.txt test.py (base) root@PC1:/home/test2# cat result.txt ## 程序运行 结果 >NC_000964.3 Bacillus subtilis subsp. subtilis str. 168 chromosome, complete genome CATTTTTTCGGCTTTTTTTAGTATCCACAGAGGTTATCGACAACATTTTCACATTACCAACCCCTGTGGACAAGGTTTTTTCAACAGGTTGTCCGCTTTGTGGATAAGATGGTGACAGCCATTGCAAGCTCTCGTTTATTTTGGTATTATATTTGTGTTTTAACTCTTGATTACTAATCCTACCTTTCCTCTTTATCCACAAAGTGTGGATAAGTTGTGGATTGATTTCACACAGCTTGTGTAGAAGGTTGTCCACAAGTTGTGAAATTTGTCGAAAAGCTATTTATCTACTATATTATATGTTTTCAACATTTAATGTGTACGAATGGTAAGCGCCATTTGCTCTTTTTTTGTGTTCTATAACAGAGAAAGACGCCATTTTCTAAGAAAAGGAGGGACGTGCCGGAAGATGGAAAATATATTAGACCTGTGGAACCAAGCCCTTGCTCAAATCGAAAAAAAGTTGAGCAAACCGAGTTTTGAGACTTGGATGAAGTCAACCAAAGCCCACTCACTGCAAGGCGATACATTAACAATCACGGCTCCCAATGAATTTGCCAGAGACTGGCTGGAGTCCAGATACTTGCATCTGATTGCAGATACTATATATGAATTAACCGGGGAAGAATTGAGCATTAAGTTTGTCATTCCTCAAAATCAAGATGTTGAGGACTTTATGCCGAAACCGCAAGTCAAAAAAGCGGTCAAAGAAGATACATCTGATTTTCCTCAAAATATGCTCAATCCAAAATATACTTTTGATACTTTTGTCATCGGATCTGGAAACCGATTTGCACATG
002、验证
参考:https://mp.weixin.qq.com/s?__biz=MzIxNzc1Mzk3NQ==&mid=2247491511&idx=1&sn=0d329538b90decbd8f7c9d6164cd2ae9&chksm=97f5afafa08226b9a38ddfaad521285d07fe5bae99ade4327ba6c39c274f807cf6ac6fc122e5&scene=178&cur_album_id=2403674812188688386#rd