http://codextechnicanum.blogspot.com/2013/12/embedding-python-in-c-converting-c.htmlHere's the entire .py file:
//Make some vectors containing the data static const double xarr[] = {1,2,3,4,5,6,7,8,9,10,11,12,13,14}; std::vector<double> xvec (xarr, xarr + sizeof(xarr) / sizeof(xarr[0]) ); static const double yarr[] = {0,0,1,1,0,0,2,2,0,0,1,1,0,0}; std::vector<double> yvec (yarr, yarr + sizeof(yarr) / sizeof(yarr[0]) ); //Transfer the C++ vector to a python tuple pXVec = PyTuple_New(xvec.size()); for (i = 0; i < xvec.size(); ++i) { pValue = PyFloat_FromDouble(xvec[i]); if (!pValue) { Py_DECREF(pXVec); Py_DECREF(pModule); fprintf(stderr, "Cannot convert array value "); return 1; } PyTuple_SetItem(pXVec, i, pValue); } //Transfer the other C++ vector to a python tuple pYVec = PyTuple_New(yvec.size()); for (i = 0; i < yvec.size(); ++i) { pValue = PyFloat_FromDouble(yvec[i]); if (!pValue) { Py_DECREF(pYVec); Py_DECREF(pModule); fprintf(stderr, "Cannot convert array value "); return 1; } PyTuple_SetItem(pYVec, i, pValue); // } //Set the argument tuple to contain the two input tuples PyTuple_SetItem(pArgTuple, 0, pXVec); PyTuple_SetItem(pArgTuple, 1, pYVec); //Call the python function pValue = PyObject_CallObject(pFunc, pArgTuple);
def plotStdVectors(x, y):
import numpy as np
import matplotlib.pyplot as plt
print "Printing from Python in plotStdVectors()"
print x
print y
x = np.fromiter(x, dtype = np.float)
y = np.fromiter(y, dtype = np.float)
print x
print y
plt.plot(x, y)
plt.show()
return 0
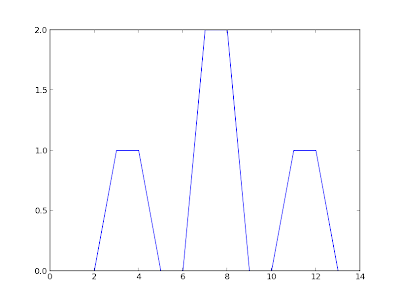
And, after compiling with the Makefile (which is for Ubuntu 12.10 using the system's default Python installation), can be run with: $ ./testEmbed pythonToEmbed plotStdVectors Hello from main Hello from runPython() Printing from Python in plotStdVectors() (1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, 10.0, 11.0, 12.0, 13.0, 14.0) (0.0, 0.0, 1.0, 1.0, 0.0, 0.0, 2.0, 2.0, 0.0, 0.0, 1.0, 1.0, 0.0, 0.0) [ 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14.] [ 0. 0. 1. 1. 0. 0. 2. 2. 0. 0. 1. 1. 0. 0.] Result of call: 0 Program finishedAnd the plot:
Real Time Plotting with C/C++ and python Concepts: Standard input/output (Think: the input and output of your program) Unix Pipes Plotting with matplotlib So, you have your code working, but you are tired of having to run a separate program to see plots? There are many simple solutions, but I am going to present what I think is the absolute simplest. What we are going to do is have your C/C++ program output the data and have python capture it and save plots in real-time! What does this entail? Well... Your C/C++ program no longer writes to a file (using fprintf), but rather writes to the standard output (STDIN, using simply printf) Your python script will now read data straight from STDIN using raw_input() You will have to use a unix shell to sew all of this together Let's look at a very simple example. Suppose we have a very important C program that outputs some very important numbers. In order for it to work with this new standard, we have it output the data to screen. Namely, it does: makedata.c #include <stdio.h> #include <math.h> #define PI 3.14159 int main() { int i,j; for(j=0; j<10; j++) { for(i=0; i<20; i++) { printf("%f ",sinf(i * PI / 10.0 + j*PI/10)); } printf(" "); } return 0; } Now, when we run this, we get a bunch of numbers thrown to screen! $ gcc -lm -o makedata makedata.c $ ./makedata 0.000000 0.309017 0.587785 0.809017 0.951056 1.000000 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 0.309017 0.587785 0.809017 0.951056 1.000000 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.587785 0.809017 0.951056 1.000000 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.809017 0.951056 1.000000 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.951056 1.000000 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.809013 1.000000 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.809013 0.951055 0.951057 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.809013 0.951055 1.000000 0.809018 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.809013 0.951055 1.000000 0.951059 0.587787 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.809013 0.951055 1.000000 0.951059 0.809021 0.309019 0.000003 -0.309014 -0.587783 -0.809015 -0.951055 -1.000000 -0.951058 -0.809020 -0.587789 -0.309022 -0.000005 0.309012 0.587781 0.809013 0.951055 1.000000 0.951059 0.809021 0.587792 In order to capture the data in python, we must use the raw_input() function. This function simply gets input from the user and puts it into a variable. It puts everything the user types up to when they press enter. This is why the C code is that it only prints a newline (ie: ' ') once one full line of data has been outputted to screen. If we had put a newline in the first printf statement, the python plotting program would only plot one number at a time! So, you can think of the tab ( ) as deliniating between values and the newline ( ) deliniating between different sets of data. The python code that reads this data looks like: plot.py import numpy as np import pylab as py def plot_data(data): py.clf() py.plot(data) py.show() py.savefig("data-%.8d.png"%counter) if __name__ == "__main__": counter = 0 while True: try: tmp = raw_input().strip().split() data = np.array(tmp, dtype=np.double) except EOFError: print "Input has terminated! Exiting" exit() except ValueError: print "Invalid input, skipping. Input was: %s"%tmp continue print "Plotting plot number %d"%counter plot_data(data) counter += 1 You can test this program by running it, typing a bunch of numbers separated by a space, then pressing enter. It will plot it, display it and save it! Then, the program will ask you again for more numbers. To exit, you type Control-D which makes the EOFError happen. What is going on in this program is quite simple. First, "tmp" gets the long string of characters that you typed in. However, python doesn't know it contains numbers, it just looks like a bunch of random characters! Now, we use numpy and tell it to create an array out of the data. The "dtype=np.double" is us telling numpy that we are realing with valid numbers. A ValueError happens if we weren't good on our promise and the input isn't in fact all numbers. Now for the most important part... how do we put these two things together? Unix has a very cool thing called input/output redirection. This allows us to redirect the output of one program to the input of another. So, instead of us having to type in the numbers for the python script, we can have the C/C++ program type it for us! The syntax is quite simple, all you have to do is: $ ./makedata | python plot.py And now you are done! You should have a bunch of plots coming up of sin waves with various phases. Congrats! There is one more thing you can do to make your plots even more fancy. Sometimes, you don't want to save each figure or have to click through to see every plot, one at a time. Instead, you just want to see an animation of what is happening as it is happening! Or, you are already making an animation with many py.plot() statements, and you want it to be smoother and faster! To do this, you can to look into pylab animations. The people at scipy have a great tutorial on this issue. You can also look at a small plotting script I made which does something very similar. The basics of this method involve: creating your plots at the beginning of your script, and saving them into variables. Then, when you get new data that you want to plot, you simply change the data in the plot with .set_data(). One thing to note is the line "py.ion()" right after I imported pylab and how I use py.draw() instead of py.show(). If you want to get started playing around with this, simply take the same code from earlier in this document, add "py.ion()" after we import pylab, delete the py.savefig() line and replace py.show() with py.draw()! This will give you a (quite slow) animation.
DECREF
http://stackoverflow.com/questions/6977161/where-should-i-put-py-incref-and-py-decref-on-this-block-in-python-c-extension
The objects you create with PyInt_FromLong() and you add to the list should be kept in a local variable.
The reason are the ownership rules: PyInt_FromLong() generates a reference that you own. In the call to PyTuple_SetItem(), you lose this ownership again, because PyTuple_SetItem() "steals" it from you, so you don't have to care about. But PyList_Append() doesn't do so, it increases the refcount. In order to have the object GC'ed correctly, you have to release your ownership by DECREF'ing.
So, instead of PyList_Append(item, PyInt_FromLong(jp)), you do the following:
PyObject * jpo = PyInt_FromLong(jp);
// do some error checking here
PyList_Append(item, jpo);
Py_DECREF(jpo);boost.python
https://www.youtube.com/watch?v=GE8EsGUsC2w
python c api
pyerr_setstring
boost
register_exception_translator
handy debugging tip
boost::python::throw_error_already_set(), set breakpoint here
https://bitbucket.org/sixty-north/scipy2014_boost_python_workshop_student_material/src
simply expose c++ function to python
boost::python::def