#数据准备
loc<-"https://archive.ics.uci.edu/ml/machine-learning-databases/"
ds<-"breast-cancer-wisconsin/breast-cancer-wisconsin.data"
url<-paste(loc,ds,sep="")
breast<-read.table(url,sep=",",header=FALSE,na.strings="?")
names(breast)<-c("ID","clumpThickness","sizeUniformity","shapeUniformity","maginalAdhesion","singleEpithelialCellSize","bareNuclei","blandChromatin","normalNucleoli","mitosis","class")
df<-breast[-1]
df$class<-factor(df$class,levels=c(2,4),labels=c("benign","malignant"))
set.seed(1234)
train<-sample(nrow(df),0.7*nrow(df))
df.train<-df[train,] #取行的意思
df.validate<-df[-train,]
table(df.train$class)
benign malignant
329 160
table(df.validate$class)
benign malignant
129 81
#逻辑回归 fit.logit<-glm(class~.,data=df.train,family=binomial()) summary(fit.logit)
Call:
glm(formula = class ~ ., family = binomial(), data = df.train)
Deviance Residuals:
Min 1Q Median 3Q Max
-2.75813 -0.10602 -0.05679 0.01237 2.64317
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -10.42758 1.47602 -7.065 1.61e-12 ***
clumpThickness 0.52434 0.15950 3.287 0.00101 **
sizeUniformity -0.04805 0.25706 -0.187 0.85171
shapeUniformity 0.42309 0.26775 1.580 0.11407
maginalAdhesion 0.29245 0.14690 1.991 0.04650 *
singleEpithelialCellSize 0.11053 0.17980 0.615 0.53871
bareNuclei 0.33570 0.10715 3.133 0.00173 **
blandChromatin 0.42353 0.20673 2.049 0.04049 *
normalNucleoli 0.28888 0.13995 2.064 0.03900 *
mitosis 0.69057 0.39829 1.734 0.08295 .
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 612.063 on 482 degrees of freedom
Residual deviance: 71.346 on 473 degrees of freedom
(6 observations deleted due to missingness)
AIC: 91.346
Number of Fisher Scoring iterations: 8
prob<-predict(fit.logit,df.validate,type="response")
logit.pred<-factor(prob>.5,levels=c(FALSE,TRUE),labels=c("begin","malignant"))
logit.perf<-table(df.validate$class,logit.pred,dnn=c("Actual","Predicted"))
logit.perf
Predicted
Actual begin malignant
benign 118 2
malignant 4 76
#决策树 library(rpart) set.seed(1234) dtree<-rpart(class~.,data=df.train,method="class",parms=list(split="information")) dtree$cptable
CP(复杂度) nsplit(树枝大小) rel error(误差) xerror(10折交叉验证误差) xstd(交叉误差的标准差)
1 0.800000 0 1.00000 1.00000 0.06484605
2 0.046875 1 0.20000 0.30625 0.04150018
3 0.012500 3 0.10625 0.20625 0.03467089
4 0.010000 4 0.09375 0.18125 0.03264401
dtree.pruned<-prune(dtree,cp=.0125) #剪枝操作 library(rpart.plot) prp(dtree.pruned,type=2,extra=104,fallen.leaves = TRUE,main="Decision Tree")

dtree.pred<-predict(dtree.pruned,df.validate,type="class")
dtree.perf<-table(df.validate$class,dtree.pred,dnn=c("Actual","Predicted"))
dtree.perf
Predicted
Actual benign malignant
benign 122 7
malignant 2 79
#条件推断树
install.packages("party")
library(party)
fit.ctree<-ctree(class~.,data=df.train)
plot(fit.ctree,main="Conditional Inference Tree")
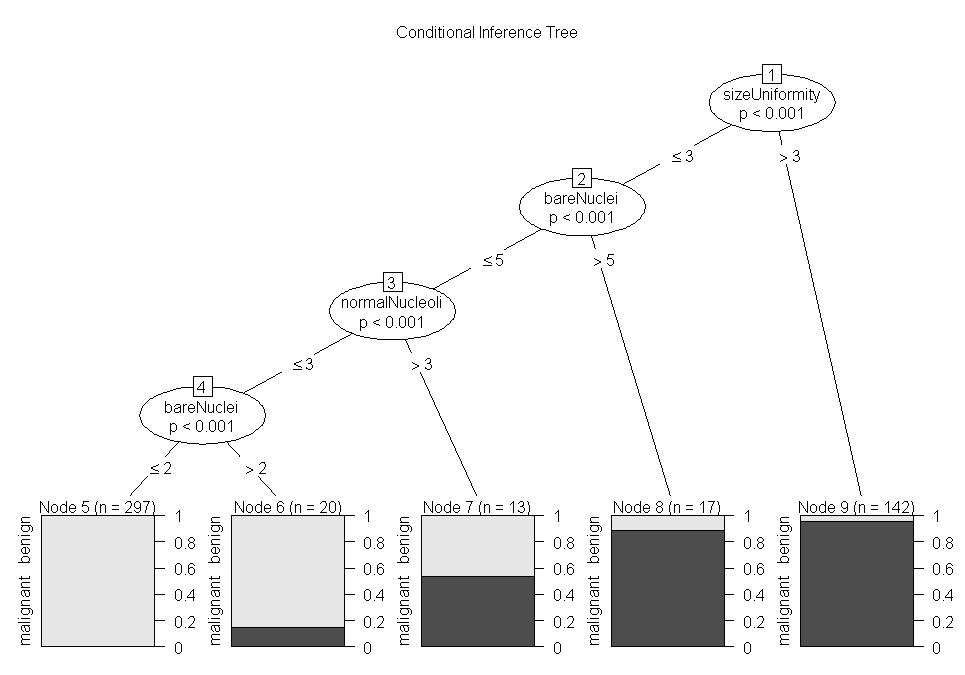
ctree.pred<-predict(fit.ctree,df.validate,type="response")
ctree.perf<-table(df.validate$class,ctree.pred,dnn=c("Actual","Predicted"))
ctree.perf
Predicted
Actual benign malignant
benign 122 7
malignant 3 78
#随机森林
install.packages("randomForest")
library(randomForest)
set.seed(1234)
fit.forest<-randomForest(class~.,data=df.train,na.action=na.roughfix,importance=TRUE)
fit.forest
Call:
randomForest(formula = class ~ ., data = df.train, importance = TRUE, na.action = na.roughfix)
Type of random forest: classification
Number of trees: 500
No. of variables tried at each split: 3
OOB estimate of error rate: 3.68%
Confusion matrix:
benign malignant class.error
benign 319 10 0.03039514
malignant 8 152 0.05000000
importance(fit.forest,type=2) #输出变量的重要性
forest.pred<-predict(fit.forest,df.validate)
forest.perf<-table(df.validate$class,forest.pred,dnn=c("Actual","Predicted"))
forest.perf
Predicted
Actual benign malignant
benign 117 3
malignant 1 79
#支持向量机
install.packages("e1071")
library(e1071)
set.seed(1234)
fit.svm<-svm(class~.,data=df.train)
fit.svm
Call:
svm(formula = class ~ ., data = df.train)
Parameters:
SVM-Type: C-classification
SVM-Kernel: radial
cost: 1
gamma: 0.1111111
Number of Support Vectors: 76
svm.pred<-predict(fit.svm,na.omit(df.validate))
svm.perf<-table(na.omit(df.validate)$class,svm.pred,dnn=c("Actual","Predicted"))
svm.perf
Predicted
Actual benign malignant
benign 116 4
malignant 3 77
#带RBF核的SVM模型 set.seed(1234) tuned<-tune.svm(class~.,data=df.train,gamma=10^(-6:1),cost=10^(-10:10)) tuned
Parameter tuning of ‘svm’:
- sampling method: 10-fold cross validation
- best parameters:
gamma cost
0.01 1
- best performance: 0.02904092
fit.svm<-svm(class~.,data=df.train,gamma=.01,cost=1)
svm.pred<-predict(fit.svm,na.omit(df.validate))
svm.perf<-table(na.omit(df.validate)$class,svm.pred,dnn=c("Actual","Predicted"))
svm.perf
Predicted
Actual benign malignant
benign 117 3
malignant 3 77
#选择预测效果最好的解,评估二分类准确性 performance<-function(table,n=2){ if(!all(dim(table)==c(2,2))) stop("Must be a 2x2 table") tn=table[1,1] fp=table[1,2] fn=table[2,1] tp=table[2,2] sensitivity=tp/(tp+fn) specificity=tn/(tn+fp) ppp=tp/(tp+fp) npp=tn/(tn+fn) hitrate=(tp+tn)/(tp+tn+fp+fn) result<-paste("Sensitivity=",round(sensitivity,n),"\nSpecificity = ",round(specificity,n),"\nPositive Predictive Value=",round(ppp,n),"\nNegative Predictive Value=",round(npp,n),"\nAccuracy=",round(hitrate,n),"\n",sep="") cat(result) } performance(logit.perf)
Sensitivity=0.95
Specificity = 0.98
Positive Predictive Value=0.97
Negative Predictive Value=0.97
Accuracy=0.97
performance(dtree.perf)
Sensitivity=0.98
Specificity = 0.95
Positive Predictive Value=0.92
Negative Predictive Value=0.98
Accuracy=0.96
performance(ctree.perf)
Sensitivity=0.96
Specificity = 0.95
Positive Predictive Value=0.92
Negative Predictive Value=0.98
Accuracy=0.95
performance(ctree.perf)
Sensitivity=0.96
Specificity = 0.95
Positive Predictive Value=0.92
Negative Predictive Value=0.98
Accuracy=0.95
performance(forest.perf)
Sensitivity=0.99
Specificity = 0.98
Positive Predictive Value=0.96
Negative Predictive Value=0.99
Accuracy=0.98
performance(svm.perf)
Sensitivity=0.96
Specificity = 0.98
Positive Predictive Value=0.96
Negative Predictive Value=0.98
Accuracy=0.97
#Rattle包
library(rattle)
loc<-"https://archive.ics.uci.edu/ml/machine-learning-databases/"
ds<-"pima-indians-diabetes/pima-indians-diabetes.data"
url<-paste(loc,ds,sep="")
diabetes<-read.table(url,sep=",",header=FALSE)
names(diabetes)<-c("npregant","plasma","bp","triceps","insulin","bmi","pedigree","age","class")
diabetes$class<-factor(diabetes$class,levels=c(0,1),labels=c("normal","diabetic"))
rattle()
cv<-matrix(c(145,50,8,27),nrow=2) performance(as.table(cv))