1.编码
这篇文章中遗传算法对TSP问题的解空间编码是十进制编码。如果有十个城市,编码可以如下:
0 1 2 3 4 5 6 7 8 9
这条编码代表着一条路径,先经过0,再经过1,依次下去。
2.选择
选择操作仍然是轮盘赌模型,虽然不会出现路径长度为负数的情况,但是需要考虑与上篇文章不同的是求的是最小值。因此在代码中概率的计算为:
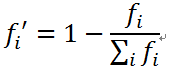
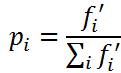
3.交叉

4.变异
变异操作就是交换两个城市,例如:
0 1 2 3 4
0 2 1 3 4
5.代码实现
#include<stdio.h> #include<string.h> #include<stdlib.h> #include<math.h> #include<time.h> #include<Windows.h> #include<fstream> #include<iostream> using namespace std; const int cities = 10; //城市的个数 const int MAXX = 100; //迭代次数 const double pc = 0.8; //交配概率 const double pm = 0.05; //变异概率 const int num = 10; //种群的大小 int bestsolution;//最优染色体 double distance1[cities][cities];//城市之间的距离 struct group //染色体的结构 { int city[cities];//城市的顺序 double adapt;//适应度 double p;//在种群中的幸存概率 }group[num],grouptemp[num]; struct point{ double x,y; }Cpoint[cities]; //随机产生cities个城市之间的相互距离 void init() {/* int i,j; memset(distance,0,sizeof(distance)); srand((unsigned)time(NULL)); for(i=0;i<cities;i++) { for(j=i+1;j<cities;j++) { distance[i][j]=rand()%100; distance[j][i]=distance[i][j]; } } //打印距离矩阵 printf("城市的距离矩阵如下 "); for(i=0;i<cities;i++) { for(j=0;j<cities;j++) printf("%4d",distance[i][j]); printf(" "); }*/ ifstream f1("C:\city.txt",ios::in); for(int i = 0; i < cities; i++){ //if(ll % 2 != 0) f1 >> Cpoint[i].x; //if(ll % 2 == 0) f1 >> Cpoint[i].y; //ll++; } f1.close(); double tmp1 = 0.0; for(int m = 0; m < cities; m++){ for(int n = 0; n < cities; n++){ if(m == n) distance1[m][n] = 0; else{ tmp1 = sqrt(pow(Cpoint[m].x - Cpoint[n].x,2) + pow(Cpoint[m].y - Cpoint[n].y,2)); distance1[m][n] = tmp1; distance1[n][m] = tmp1; } } } //打印距离矩阵 printf("城市的距离矩阵如下 "); for(int i = 0; i < cities; i++) { for(int j = 0; j<cities; j++){ cout << distance1[i][j] << ' '; } cout << endl; } } //随机产生初试群 void groupproduce() { int i,j,t,k,flag; for(i = 0; i < num; i++) //初始化 for(j = 0; j < cities; j++) group[i].city[j]=-1; srand((unsigned)time(NULL)); for(i = 0; i < num; i++) { //产生10个不相同的数字 for(j = 0; j < cities;) { t = rand() % cities; flag = 1; for(k = 0; k < j; k++) { if(group[i].city[k] == t) { flag = 0; break; } } if(flag) { group[i].city[j] = t; j++; } } } //打印种群基因 printf("初始的种群 "); for(i = 0; i < num; i++) { for(j = 0; j < cities; j++) printf("%4d",group[i].city[j]); printf(" "); } } //评价函数,找出最优染色体 void pingjia() { int i,j; int n1,n2; double sumdistance; int biggestsum = 0; double biggestp = 0; for(i = 0; i < num; i++) { sumdistance = 0; for(j = 1; j < cities; j++) { n1 = group[i].city[j-1]; n2 = group[i].city[j]; sumdistance += distance1[n1][n2]; } sumdistance += distance1[group[i].city[cities-1]][group[i].city[0]]; group[i].adapt = sumdistance; //每条染色体的路径总和 //printf("%d",group[i].adapt); biggestsum += sumdistance; //种群的总路径 } //计算染色体的幸存能力,路劲越短生存概率越大 for(i = 0; i < num; i++) { group[i].p = 1 - (double)group[i].adapt / (double)biggestsum; biggestp += group[i].p; //printf("%s"," "); } //printf("%f",biggestp); for(i = 0; i < num; i++){ group[i].p = group[i].p / biggestp; }//在种群中的幸存概率,总和为1 //求最佳路径 bestsolution = 0; for(i = 0;i < num; i++) if(group[i].p > group[bestsolution].p) bestsolution = i; //打印适应度 /*for(i = 0; i < num; i++) cout << "染色体" << i << "的路径之和与生存概率分别为" << group[i].adapt << "和" << group[i].p; //printf("染色体%d的路径之和与生存概率分别为%4d %.4f ",i,group[i].adapt,group[i].p); cout << "当前种群的最优染色体是" << bestsolution << "号染色体" << endl; //printf("当前种群的最优染色体是%d号染色体 ",bestsolution);*/ } //选择 void xuanze() { int i,j,temp; double gradient[num];//梯度概率 double xuanze[num];//选择染色体的随机概率 int xuan[num];//选择了的染色体 //初始化梯度概率 for(i = 0; i < num; i++) { gradient[i] = 0.0; xuanze[i] = 0.0; } gradient[0] = group[0].p; for(i = 1; i < num; i++) gradient[i] = gradient[i-1]+group[i].p; srand((unsigned)time(NULL)); //随机产生染色体的存活概率 for(i = 0; i < num; i++) { xuanze[i] = (rand()%100); xuanze[i] /= 100; } //选择能生存的染色体 for( i = 0; i < num; i++) { for(j = 0; j < num; j++) { if(xuanze[i] < gradient[j]) { xuan[i] = j; //第i个位置存放第j个染色体 break; } } } //拷贝种群 for(i = 0; i < num; i++) { grouptemp[i].adapt = group[i].adapt; grouptemp[i].p = group[i].p; for(j = 0; j < cities; j++) grouptemp[i].city[j] = group[i].city[j]; } //数据更新 for(i = 0; i < num; i++) { temp = xuan[i]; group[i].adapt = grouptemp[temp].adapt; group[i].p = grouptemp[temp].p; for(j = 0; j < cities; j++) group[i].city[j] = grouptemp[temp].city[j]; } //用于测试 /* printf("<-------------------------------> "); for(i=0;i<num;i++) { for(j=0;j<cities;j++) printf("%4d",group[i].city[j]); printf(" "); printf("染色体%d的路径之和与生存概率分别为%4d %.4f ",i,group[i].adapt,group[i].p); } */ } //交配,对每个染色体产生交配概率,满足交配率的染色体进行交配 void jiaopei() { int i,j,k,kk; int t;//参与交配的染色体的个数 int point1,point2,temp;//交配断点 int pointnum; int temp1,temp2; int map1[cities],map2[cities]; double jiaopeip[num];//染色体的交配概率 int jiaopeiflag[num];//染色体的可交配情况 int kkk,flag=0; //初始化 for(i = 0; i < num; i++) { jiaopeiflag[i] = 0; } //随机产生交配概率 srand((unsigned)time(NULL)); for(i = 0; i < num; i++) { jiaopeip[i] = (rand()%100); jiaopeip[i] /= 100; } //确定可以交配的染色体 t = 0; for(i = 0; i < num; i++) { if(jiaopeip[i] < pc) { jiaopeiflag[i] = 1; t++; } } t = t/2 * 2;//t必须为偶数,产生t/2个0-9交配断点 srand((unsigned)time(NULL)); temp1 = 0; //temp1号染色体和temp2染色体交配 for(i = 0; i < t/2; i++) //如果有5个染色体需要交配,但是实际上t/2代表只有4个染色体会真正的交配,剩下的1个再加上5个不需要交配的染色体直接进入下一代。 { point1 = rand() % cities;//交配点1 point2 = rand() % cities;//交配点2 //选出一个需要交配的染色体1 for(j = temp1;j < num; j++) { if(jiaopeiflag[j] == 1) { temp1 = j; break; } } //选出另一个需要交配的染色体2与1交配 for(j = temp1+1; j < num; j++) { if(jiaopeiflag[j] == 1) { temp2 = j; break; } } //进行基因交配 if(point1 > point2) //保证point1<=point2 { temp = point1; point1 = point2; point2 = temp; } //初始化 memset(map1,-1,sizeof(map1)); memset(map2,-1,sizeof(map2)); //断点之间的基因产生映射 for(k = point1; k <= point2; k++) { map1[group[temp1].city[k]] = group[temp2].city[k]; map2[group[temp2].city[k]] = group[temp1].city[k]; } //断点两边的基因互换 for(k = 0; k < point1; k++) { temp = group[temp1].city[k]; group[temp1].city[k] = group[temp2].city[k]; group[temp2].city[k] = temp; } for(k = point2+1; k < cities; k++) { temp = group[temp1].city[k]; group[temp1].city[k] = group[temp2].city[k]; group[temp2].city[k] = temp; } //printf("处理冲突--------------------- "); //处理染色体1产生的冲突基因 for(k = 0; k < point1; k++) { for(kk = point1; kk <= point2; kk++) { if(group[temp1].city[k] == group[temp1].city[kk]) { group[temp1].city[k] = map1[group[temp1].city[k]]; //如果相等则进行映射操作 //find for(kkk = point1;kkk <= point2; kkk++) { if(group[temp1].city[k] == group[temp1].city[kkk]) //考虑如果映射一次仍然具有相同的城市,则再进行一次映射操作 { flag = 1; break; } } if(flag == 1) //flag不断判断同一染色体中是否还存在相同的城市 { kk = point1 - 1; flag = 0; } else { flag = 0; break; } } } } for(k = point2+1; k < cities; k++) { for(kk = point1; kk <= point2; kk++) { if(group[temp1].city[k] == group[temp1].city[kk]) { group[temp1].city[k] = map1[group[temp1].city[k]]; //find for(kkk = point1;kkk <= point2; kkk++) { if(group[temp1].city[k] == group[temp1].city[kkk]) { flag = 1; break; } } if(flag == 1) { kk = point1 - 1; flag = 0; } else { flag = 0; break; } } } } //处理2染色体产生的冲突基因 for(k = 0;k < point1; k++) { for(kk = point1; kk <= point2; kk++) { if(group[temp2].city[k] == group[temp2].city[kk]) { group[temp2].city[k] = map2[group[temp2].city[k]]; //find for(kkk = point1;kkk <= point2; kkk++) { if(group[temp2].city[k] == group[temp2].city[kkk]) { flag = 1; break; } } if(flag == 1) { kk = point1 - 1; flag = 0; } else { flag = 0; break; } } } } for(k = point2+1; k < cities; k++) { for(kk = point1; kk <= point2; kk++) { if(group[temp2].city[k] == group[temp2].city[kk]) { group[temp2].city[k] = map2[group[temp2].city[k]]; //find for(kkk = point1; kkk <= point2; kkk++) { if(group[temp2].city[k] == group[temp2].city[kkk]) { flag = 1; break; } } if(flag == 1) { kk = point1 - 1; flag = 0; } else { flag = 0; break; } } } } temp1 = temp2 + 1; } } void bianyi() { int i,j; int t; int temp1,temp2,point; double bianyip[num]; //染色体的变异概率 int bianyiflag[num];//染色体的变异情况 for(i = 0;i < num; i++)//初始化 bianyiflag[i]=0; //随机产生变异概率 srand((unsigned)time(NULL)); for(i = 0; i < num; i++) { bianyip[i] = (rand()%100); bianyip[i] /= 100; } //确定可以变异的染色体 t=0; for(i = 0; i < num; i++) { if(bianyip[i] < pm) { bianyiflag[i] = 1; t++; } } //变异操作,即交换染色体的两个节点 srand((unsigned)time(NULL)); for(i = 0; i < num; i++) { if(bianyiflag[i] == 1) { temp1 = rand() % 10; temp2 = rand() % 10; point = group[i].city[temp1]; group[i].city[temp1] = group[i].city[temp2]; group[i].city[temp2] = point; } } } int main() { int i,j,t; init(); groupproduce(); //初始种群评价 pingjia(); t=0; while(t++ < MAXX) { xuanze(); jiaopei(); bianyi(); pingjia(); } //最终种群的评价 printf(" 输出最终的种群评价 "); for(i = 0; i < num; i++) { for(j = 0;j < cities; j++) { printf("%4d",group[i].city[j]); } cout << " adapt:" << group[i].adapt << " p:" << group[i].p << endl; //printf(" adapt:%4d, p:%.4f ",group[i].adapt,group[i].p); } printf("最优解为%d号染色体 ",bestsolution); system("pause"); return 0; }
6.结果显示
我的城市为10个,坐标为
41 94
37 84
53 67
25 62
7 64
2 99
68 58
71 44
54 62
83 69
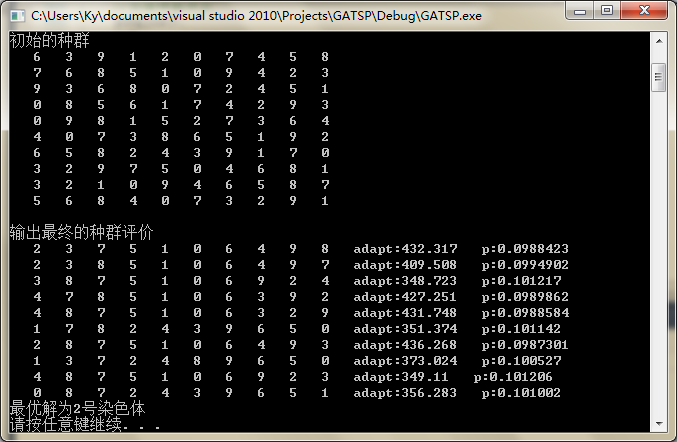
输出结果如图:
7.参考
1.http://blog.csdn.net/mylovestart/article/details/8977005#cpp
2.http://blog.csdn.net/yeruby/article/details/13161853