InterProScan是一个蛋白质功能数据库,输入蛋白质结构和位点可以预测蛋白质功能。
interproscan回答的问题是:I have a new protein sequence, and I don’t know anything about it. Is there some known motif in it that would help me assign a function to the protein?
如果手里有一条不知道有何作用的蛋白质,我能否搜索一些当中的motif用来鉴定它的function?
motif就是protein的二级结构,interproscan拿到protein sequence之后,在member database中搜索二级结构,这样做的结果是得到了很多二级结构的annotation,然后将这些annotation 整合起来得到一个完整的protein structure,同时,这个motif在interproscan中也有一个编号就是IPR编号,它可能与PF编号所代表的motif相同
for example, the same motif is known as PF01623 in Pfam and as IPR002568 in InterPro.
eg:

For example, for the motif IPR002568, the GO term GO:0003676 is returned. This term means that the found motif is related to the nucleic acid binding function.
Interproscan,通过蛋白质结构域和功能位点数据库预测蛋白质功能。是EBI开发的一个集成了蛋白质家族、结构域和功能位点的非冗余数据库。Interproscan整合了一些使用最普及的一些数据库,并应用于功能未知的蛋白进行Interpro注释和GO注释。
Proteins that have diverged from a common ancestral gene are known as homologous,所以homologous就是祖先gene

analysis:protein的几种分析切入点:1.基于domain2.基于sequence feature(也就是function)
gene family中的gene function related且来自同一个ancestor gene,gene family的classification based on their diversity and function,A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions.也就是说,首先check是否有相同homologous的duplication(也就是based on domain) 然后再check their own function(based on sequence feature)。
Although genes differ in sequence, size, and functional domains, they can be grouped into families based on their homology
domain是一个protein的组成部分,同一个protein中含有不同的domain

这是一个protein Nck,其中SH3和SH2都是domain,这个protein Nck由3个SH3和一个SH2(scr homologous 2)组成,不同的domain具有不同功能,比如:
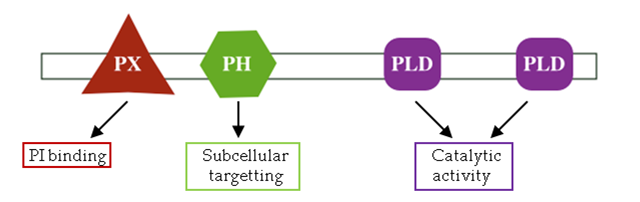
这些具有不同功能的domains,将它们的功能assembly,一同完成一个大的行为

同一个gene family中的gene,比如RGS1、RGS3和RGS6中都有相同的domain

sequence feature包括了
active site(激活位点)就是酶作用位点,酶作用之后便free了
In biology, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of residues that form temporary bonds with the substrate (binding site) and residues that catalyse a reaction of that substrate (catalytic site).
binding site是residue binding site
Active sites are present in enzymes. It is the site where the substrate binds and product is formed. And the enzyme is free for another substrate binding after product is formed. Binding sites are where any residue binds, no reaction or product formation occurs here
PTM含有化学修饰位点
repeat:sequence repeat region
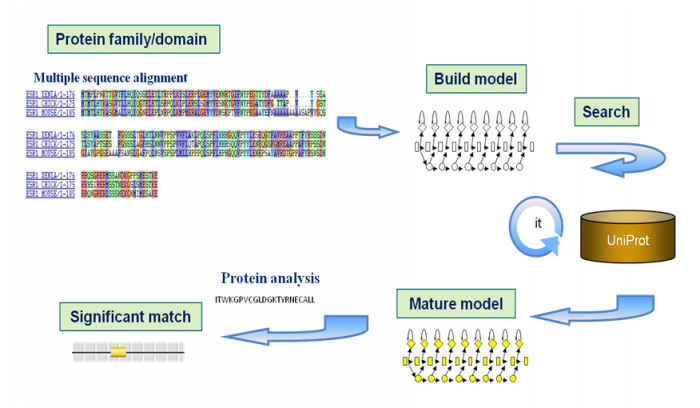
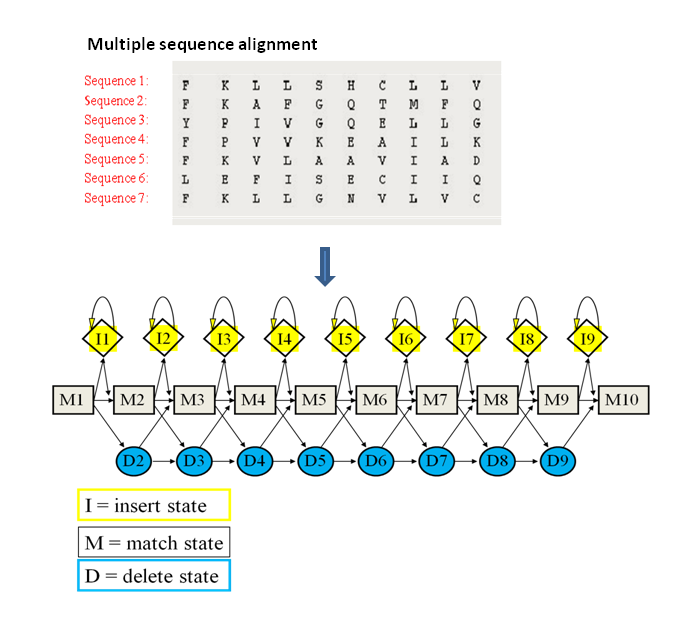
首先 mutilply sequence alignment找到相同structure,可以认为是ancestor gene(如下图中选出两个残基,这两个残基在所有物种中都存在,所以认为是比较保守的),然后built models,这仅是一个initial model,此时需要put initial model into the model databse,在database中search same model,得到的model related to the intial model 就是mature model(这个就是protein signature),最后做analysis。
One set of such tools are the predictive models known as protein signatures.
Active sites are present in enzymes. It is the site where the substrate binds and product is formed. And the enzyme is free for another substrate binding after product is formed. Binding sites are where any residue binds, no reaction or product formation occurs here
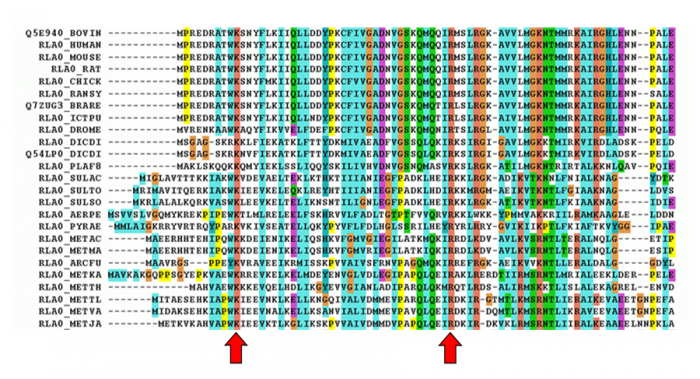
比较多个protein signature是一个process,start from the multiple sequence alignment
patterns

pattern就是现象抽取出来的数学表达,如上图中的regular expression:
When creating patterns, a conserved motif is used to build a regular expression.
The pattern illustrated here is translated as: [Ala or Cys]-any-Val-any-any-any-any-{any but Glu or Asp}.
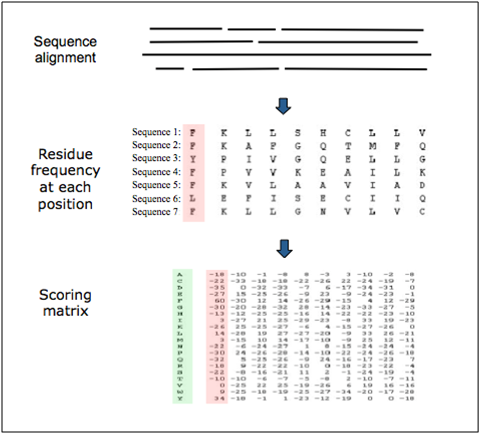
Representation of a scoring matrix based on a multiple sequence alignment. Each of the 20 amino acids commonly found in proteins is given a score for each position in the sequence according to the frequency with which they occur in the original alignment. Other factors, such as evolutionary distances can also be considered.
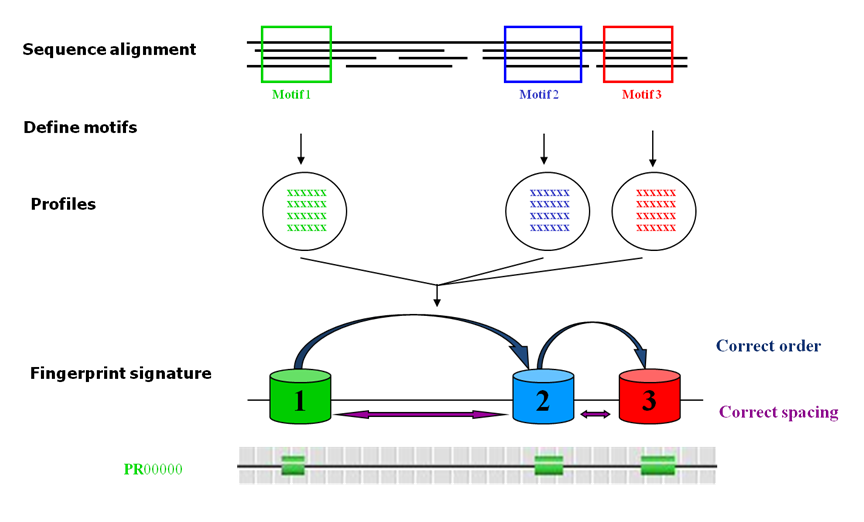
fingerprints
hidden Markov models (HMMs)