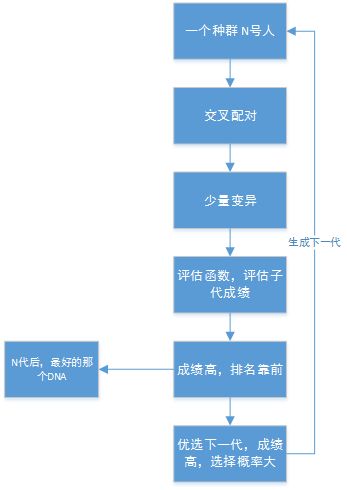
遗传算法:
先上代码:
# -*- coding: UTF-8 -*- """ Visualize Genetic Algorithm to match the target phrase. Visit my tutorial website for more: https://morvanzhou.github.io/tutorials/ """ import numpy as np TARGET_PHRASE = 'You get it!' # target DNA 最后目标 POP_SIZE = 300 # population size 300 个祖宗 CROSS_RATE = 0.4 # mating probability (DNA crossover) MUTATION_RATE = 0.01 # mutation probability 变异 N_GENERATIONS = 1000 #进化代数 DNA_SIZE = len(TARGET_PHRASE) TARGET_ASCII = np.fromstring(TARGET_PHRASE, dtype=np.uint8) # convert string to number ASCII_BOUND = [32, 126] # ascii 范围 class GA(object): def __init__(self, DNA_size, DNA_bound, cross_rate, mutation_rate, pop_size): self.DNA_size = DNA_size DNA_bound[1] += 1 self.DNA_bound = DNA_bound self.cross_rate = cross_rate self.mutate_rate = mutation_rate self.pop_size = pop_size # 生成第一代 300个祖宗 , 数据类型 (300,11) =( pop_size, dna_size) 32 ~ 128 之间 self.pop = np.random.randint(*DNA_bound, size=(pop_size, DNA_size)).astype(np.int8) # int8 for convert to ASCII def translateDNA(self, DNA): # convert to readable string return DNA.tostring().decode('ascii') def get_fitness(self): # count how many character matches match_count = (self.pop == TARGET_ASCII).sum(axis=1) return match_count def select(self): #排序生成输出, 有优势的 被选出概率更高 fitness = self.get_fitness() + 1e-4 # add a small amount to avoid all zero fitness idx = np.random.choice(np.arange(self.pop_size), size=self.pop_size, replace=True, p=fitness/fitness.sum()) ''' data = fitness/fitness.sum() print data print idx print data.shape print idx.shape ''' return self.pop[idx] def crossover(self, parent, pop): #交叉生娃: 某一个男人,去随机选一个女人,交叉配对部分数据; 男人是否有权利靠 np.random.rand() < self.cross_rate if np.random.rand() < self.cross_rate: i_ = np.random.randint(0, self.pop_size, size=1) # select another individual from pop cross_points = np.random.randint(0, 2, self.DNA_size).astype(np.bool) # choose crossover points parent[cross_points] = pop[i_, cross_points] # mating and produce one child #print cross_points 50% 交叉 return parent def mutate(self, child): #小变异 for point in range(self.DNA_size): if np.random.rand() < self.mutate_rate: child[point] = np.random.randint(*self.DNA_bound) # choose a random ASCII index return child def evolve(self): # 进化 pop = self.select() #print pop.shape pop_copy = pop.copy() #print 'pop_copy:', pop_copy for parent in pop: # for every parent child = self.crossover(parent, pop_copy) child = self.mutate(child) parent[:] = child #print 'child:',child #print 'parent:',parent self.pop = pop if __name__ == '__main__': ''' 300 个祖宗 # np.random.randint(*DNA_bound, size=(pop_size, DNA_size)).astype(np.int8) data = np.random.randint(*ASCII_BOUND, size=(POP_SIZE, DNA_SIZE)) print data # (POP_SIZE, DNA_SIZE)).astype(np.int8) print data.shape data2 = data.astype(np.int8) print data2 print data2.shape exit() ''' ga = GA(DNA_size=DNA_SIZE, DNA_bound=ASCII_BOUND, cross_rate=CROSS_RATE, mutation_rate=MUTATION_RATE, pop_size=POP_SIZE) #ga.select( ) ''' print ga.get_fitness() eqlist = (ga.pop == TARGET_ASCII) print eqlist #.sum(axis=1) print eqlist.shape eqlistsum = eqlist.sum(axis=1) print eqlistsum #.sum(axis=1) print eqlistsum.shape exit() ''' for generation in range(N_GENERATIONS): fitness = ga.get_fitness() best_DNA = ga.pop[ np.argmax(fitness) ] best_phrase = ga.translateDNA(best_DNA) print('Gen', generation, ': ', best_phrase) if best_phrase == TARGET_PHRASE: break ga.evolve()
原理理解: