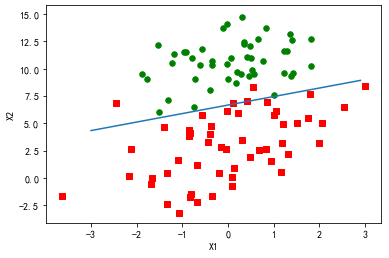
假设现在有一些数据点,我们用
一条直线对这些点进行拟合(该线称为最佳拟合直线),这个拟合过程就称作回归。利用Logistic
回归进行分类的主要思想是:根据现有数据对分类边界线建立回归公式,以此进行分类。这里的
“ 回归” 一词源于最佳拟合,表示要找到最佳拟合参数集。
训练分类器时的做法就是寻找最佳拟合参数,使用的是最优化算法。

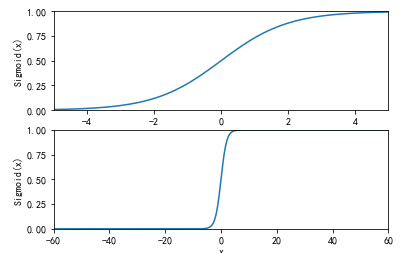
基于Logistic回归和Sigmoid函数的分类


import sys from pylab import * t = arange(-60.0, 60.3, 0.1) s = 1/(1 + exp(-t)) ax = subplot(211) ax.plot(t,s) ax.axis([-5,5,0,1]) plt.xlabel('x') plt.ylabel('Sigmoid(x)') ax = subplot(212) ax.plot(t,s) ax.axis([-60,60,0,1]) plt.xlabel('x') plt.ylabel('Sigmoid(x)') show()

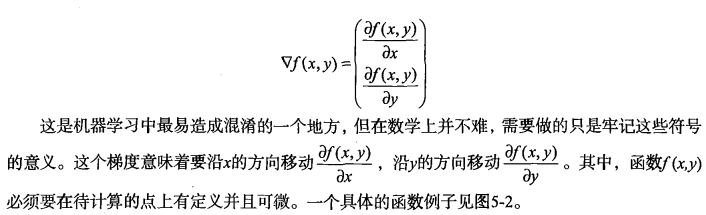
基于最优化方法的最佳回归系数确定

梯度上升法
梯度上升法基于的思想是:要找到某函数的
最大值,最好的方法是沿着该函数的梯度方向探寻。
import matplotlib import numpy as np import matplotlib.cm as cm import matplotlib.mlab as mlab import matplotlib.pyplot as plt leafNode = dict(boxstyle="round4", fc="0.8") arrow_args = dict(arrowstyle="<-") matplotlib.rcParams['xtick.direction'] = 'out' matplotlib.rcParams['ytick.direction'] = 'out' delta = 0.025 x = np.arange(-2.0, 2.0, delta) y = np.arange(-2.0, 2.0, delta) X, Y = np.meshgrid(x, y) Z1 = -((X-1)**2) Z2 = -(Y**2) #Z1 = mlab.bivariate_normal(X, Y, 1.0, 1.0, 0.0, 0.0) #Z2 = mlab.bivariate_normal(X, Y, 1.5, 0.5, 1, 1) # difference of Gaussians Z = 1.0 * (Z2 + Z1)+5.0 # Create a simple contour plot with labels using default colors. The # inline argument to clabel will control whether the labels are draw # over the line segments of the contour, removing the lines beneath # the label plt.figure() CS = plt.contour(X, Y, Z) plt.annotate('', xy=(0.05, 0.05), xycoords='axes fraction',xytext=(0.2,0.2), textcoords='axes fraction',va="center", ha="center", bbox=leafNode, arrowprops=arrow_args ) plt.text(-1.9, -1.8, 'P0') plt.annotate('', xy=(0.2,0.2), xycoords='axes fraction',xytext=(0.35,0.3), textcoords='axes fraction',va="center", ha="center", bbox=leafNode, arrowprops=arrow_args ) plt.text(-1.35, -1.23, 'P1') plt.annotate('', xy=(0.35,0.3), xycoords='axes fraction',xytext=(0.45,0.35), textcoords='axes fraction',va="center", ha="center", bbox=leafNode, arrowprops=arrow_args ) plt.text(-0.7, -0.8, 'P2') plt.text(-0.3, -0.6, 'P3') plt.clabel(CS, inline=1, fontsize=10) plt.title('Gradient Ascent') plt.xlabel('x') plt.ylabel('y') plt.show()
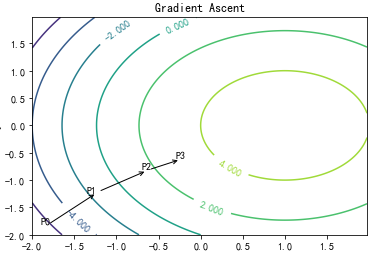

可以看到,梯度算子总是指向函数值增长
最快的方向。这里所说的是移动方向,而未提到移动量的大小。该量值称为步长,记做a用向
量来表示的话,梯度算法的迭代公式如下:
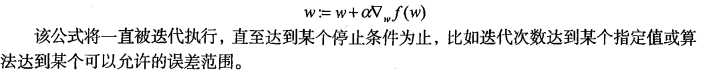

训练算法:使用梯度上升找到最佳参数

from numpy import * def loadDataSet(): dataMat = []; labelMat = [] fr = open('testSet.txt') for line in fr.readlines(): lineArr = line.strip().split() dataMat.append([1.0, float(lineArr[0]), float(lineArr[1])]) labelMat.append(int(lineArr[2])) return dataMat,labelMat def sigmoid(inX): return 1.0/(1+exp(-inX)) def gradAscent(dataMatIn, classLabels): dataMatrix = mat(dataMatIn) #convert to NumPy matrix labelMat = mat(classLabels).transpose() #convert to NumPy matrix m,n = shape(dataMatrix) alpha = 0.001 maxCycles = 500 weights = ones((n,1)) for k in range(maxCycles): #heavy on matrix operations h = sigmoid(dataMatrix*weights) #matrix mult error = (labelMat - h) #vector subtraction weights = weights + alpha * dataMatrix.transpose()* error #matrix mult return weights
dataMat,labelMat = loadDataSet() weights = gradAscent(dataMat,labelMat) print(weights)

分析数据:画出决策边界
上面已经解出了一组回归系数,它确定了不同类别数据之间的分隔线。
import matplotlib import matplotlib.pyplot as plt from numpy import * from matplotlib.patches import Rectangle def loadDataSet(): dataMat = [] labelMat = [] fr = open('F:\machinelearninginaction\Ch05\testSet.txt') for line in fr.readlines(): lineArr = line.strip().split() dataMat.append([1.0, float(lineArr[0]), float(lineArr[1])]) labelMat.append(int(lineArr[2])) return dataMat,labelMat def sigmoid(inX): return 1.0/(1+exp(-inX)) def stocGradAscent0(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.01 weights = ones(n) #initialize to all ones for i in range(m): h = sigmoid(sum(dataMatrix[i]*weights)) error = classLabels[i] - h weights = weights + alpha * error * dataMatrix[i] return weights def gradAscent(dataMatIn, classLabels): dataMatrix = mat(dataMatIn) #convert to NumPy matrix labelMat = mat(classLabels).transpose() #convert to NumPy matrix m,n = shape(dataMatrix) alpha = 0.001 maxCycles = 500 weights = ones((n,1)) for k in range(maxCycles): #heavy on matrix operations h = sigmoid(dataMatrix*weights) #matrix mult error = (labelMat - h) #vector subtraction weights = weights + alpha * dataMatrix.transpose()* error #matrix mult return weights dataMat,labelMat=loadDataSet() dataArr = array(dataMat) weights = gradAscent(dataArr,labelMat) n = shape(dataArr)[0] #number of points to create xcord1 = [] ycord1 = [] xcord2 = [] ycord2 = [] markers =[] colors =[] for i in range(n): if int(labelMat[i])== 1: xcord1.append(dataArr[i,1]); ycord1.append(dataArr[i,2]) else: xcord2.append(dataArr[i,1]); ycord2.append(dataArr[i,2]) fig = plt.figure() ax = fig.add_subplot(111) #ax.scatter(xcord,ycord, c=colors, s=markers) type1 = ax.scatter(xcord1, ycord1, s=30, c='red', marker='s') type2 = ax.scatter(xcord2, ycord2, s=30, c='green') x = arange(-3.0, 3.0, 0.1) #weights = [-2.9, 0.72, 1.29] #weights = [-5, 1.09, 1.42] weights = [13.03822793, 1.32877317, -1.96702074] weights = [4.12, 0.48, -0.6168] y = (-weights[0]-weights[1]*x)/weights[2] type3 = ax.plot(x, y) #ax.legend([type1, type2, type3], ["Did Not Like", "Liked in Small Doses", "Liked in Large Doses"], loc=2) #ax.axis([-5000,100000,-2,25]) plt.xlabel('X1') plt.ylabel('X2') plt.show()
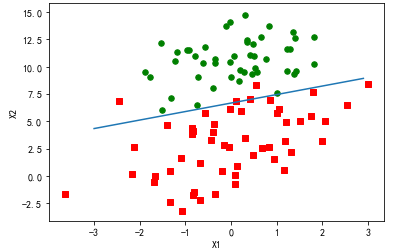
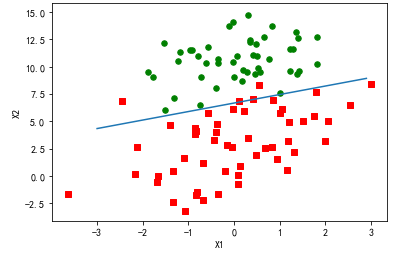
训练算法:随机梯度上升

def stocGradAscent0(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.01 weights = ones(n) #initialize to all ones for i in range(m): h = sigmoid(sum(dataMatrix[i]*weights)) error = classLabels[i] - h weights = weights + alpha * error * dataMatrix[i] return weights
import matplotlib import matplotlib.pyplot as plt from numpy import * from matplotlib.patches import Rectangle def loadDataSet(): dataMat = [] labelMat = [] fr = open('F:\machinelearninginaction\Ch05\testSet.txt') for line in fr.readlines(): lineArr = line.strip().split() dataMat.append([1.0, float(lineArr[0]), float(lineArr[1])]) labelMat.append(int(lineArr[2])) return dataMat,labelMat def sigmoid(inX): return 1.0/(1+exp(-inX)) def stocGradAscent0(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.01 weights = ones(n) #initialize to all ones for i in range(m): h = sigmoid(sum(dataMatrix[i]*weights)) error = classLabels[i] - h weights = weights + alpha * error * dataMatrix[i] return weights def gradAscent(dataMatIn, classLabels): dataMatrix = mat(dataMatIn) #convert to NumPy matrix labelMat = mat(classLabels).transpose() #convert to NumPy matrix m,n = shape(dataMatrix) alpha = 0.001 maxCycles = 500 weights = ones((n,1)) for k in range(maxCycles): #heavy on matrix operations h = sigmoid(dataMatrix*weights) #matrix mult error = (labelMat - h) #vector subtraction weights = weights + alpha * dataMatrix.transpose()* error #matrix mult return weights dataMat,labelMat=loadDataSet() dataArr = array(dataMat) weights = stocGradAscent0(dataArr,labelMat) n = shape(dataArr)[0] #number of points to create xcord1 = [] ycord1 = [] xcord2 = [] ycord2 = [] markers =[] colors =[] for i in range(n): if int(labelMat[i])== 1: xcord1.append(dataArr[i,1]); ycord1.append(dataArr[i,2]) else: xcord2.append(dataArr[i,1]); ycord2.append(dataArr[i,2]) fig = plt.figure() ax = fig.add_subplot(111) #ax.scatter(xcord,ycord, c=colors, s=markers) type1 = ax.scatter(xcord1, ycord1, s=30, c='red', marker='s') type2 = ax.scatter(xcord2, ycord2, s=30, c='green') x = arange(-3.0, 3.0, 0.1) #weights = [-2.9, 0.72, 1.29] #weights = [-5, 1.09, 1.42] weights = [13.03822793, 1.32877317, -1.96702074] weights = [4.12, 0.48, -0.6168] y = (-weights[0]-weights[1]*x)/weights[2] type3 = ax.plot(x, y) #ax.legend([type1, type2, type3], ["Did Not Like", "Liked in Small Doses", "Liked in Large Doses"], loc=2) #ax.axis([-5000,100000,-2,25]) plt.xlabel('X1') plt.ylabel('X2') plt.show()
import matplotlib import matplotlib.pyplot as plt from numpy import * from matplotlib.patches import Rectangle def loadDataSet(): dataMat = [] labelMat = [] fr = open('F:\machinelearninginaction\Ch05\testSet.txt') for line in fr.readlines(): lineArr = line.strip().split() dataMat.append([1.0, float(lineArr[0]), float(lineArr[1])]) labelMat.append(int(lineArr[2])) return dataMat,labelMat def sigmoid(inX): return 1.0/(1+exp(-inX)) def stocGradAscent0(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.5 weights = ones(n) #initialize to all ones weightsHistory=zeros((500*m,n)) for j in range(500): for i in range(m): h = sigmoid(sum(dataMatrix[i]*weights)) error = classLabels[i] - h weights = weights + alpha * error * dataMatrix[i] weightsHistory[j*m + i,:] = weights return weightsHistory def stocGradAscent1(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.4 weights = ones(n) #initialize to all ones weightsHistory=zeros((40*m,n)) for j in range(40): dataIndex = range(m) for i in range(m): alpha = 4/(1.0+j+i)+0.01 randIndex = int(random.uniform(0,len(dataIndex))) h = sigmoid(sum(dataMatrix[randIndex]*weights)) error = classLabels[randIndex] - h #print error weights = weights + alpha * error * dataMatrix[randIndex] weightsHistory[j*m + i,:] = weights # del(dataIndex[randIndex]) print(weights) return weightsHistory dataMat,labelMat=loadDataSet() dataArr = array(dataMat) myHist = stocGradAscent1(dataArr,labelMat) n = shape(dataArr)[0] #number of points to create xcord1 = [] ycord1 = [] xcord2 = [] ycord2 = [] markers =[] colors =[] fig = plt.figure() ax = fig.add_subplot(311) type1 = ax.plot(myHist[:,0]) plt.ylabel('X0') ax = fig.add_subplot(312) type1 = ax.plot(myHist[:,1]) plt.ylabel('X1') ax = fig.add_subplot(313) type1 = ax.plot(myHist[:,2]) plt.xlabel('iteration') plt.ylabel('X2') plt.show()
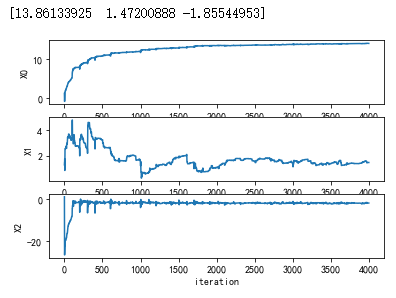

值 得注意的是,在大的波动停止后,还有一些小的周期性波动。不难理解,产生这种现象的原因是
存在一些不能正确分类的样本点(数据集并非线性可分),在每次迭代时会引发系数的剧烈改变。
我们期望算法能避免来回波动,从而收敛到某个值。另外,收敛速度也需要加快。
改进的随机梯度上升算法
def stocGradAscent1(dataMatrix, classLabels, numIter=150): m,n = shape(dataMatrix) weights = ones(n) #initialize to all ones for j in range(numIter): dataIndex = range(m) for i in range(m): alpha = 4/(1.0+j+i)+0.0001 #apha decreases with iteration, does not randIndex = int(random.uniform(0,len(dataIndex)))#go to 0 because of the constant h = sigmoid(sum(dataMatrix[randIndex]*weights)) error = classLabels[randIndex] - h weights = weights + alpha * error * dataMatrix[randIndex] del(dataIndex[randIndex]) return weights
import matplotlib import matplotlib.pyplot as plt from numpy import * from matplotlib.patches import Rectangle def loadDataSet(): dataMat = [] labelMat = [] fr = open('F:\machinelearninginaction\Ch05\testSet.txt') for line in fr.readlines(): lineArr = line.strip().split() dataMat.append([1.0, float(lineArr[0]), float(lineArr[1])]) labelMat.append(int(lineArr[2])) return dataMat,labelMat def sigmoid(inX): return 1.0/(1+exp(-inX)) def stocGradAscent0(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.5 weights = ones(n) #initialize to all ones weightsHistory=zeros((500*m,n)) for j in range(500): for i in range(m): h = sigmoid(sum(dataMatrix[i]*weights)) error = classLabels[i] - h weights = weights + alpha * error * dataMatrix[i] weightsHistory[j*m + i,:] = weights return weightsHistory def stocGradAscent1(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.4 weights = ones(n) #initialize to all ones weightsHistory=zeros((40*m,n)) for j in range(40): dataIndex = range(m) for i in range(m): alpha = 4/(1.0+j+i)+0.01 randIndex = int(random.uniform(0,len(dataIndex))) h = sigmoid(sum(dataMatrix[randIndex]*weights)) error = classLabels[randIndex] - h #print error weights = weights + alpha * error * dataMatrix[randIndex] weightsHistory[j*m + i,:] = weights # del(dataIndex[randIndex]) print(weights) return weightsHistory dataMat,labelMat=loadDataSet() dataArr = array(dataMat) myHist = stocGradAscent1(dataArr,labelMat) n = shape(dataArr)[0] #number of points to create xcord1 = [] ycord1 = [] xcord2 = [] ycord2 = [] markers =[] colors =[] fig = plt.figure() ax = fig.add_subplot(311) type1 = ax.plot(myHist[:,0]) plt.ylabel('X0') ax = fig.add_subplot(312) type1 = ax.plot(myHist[:,1]) plt.ylabel('X1') ax = fig.add_subplot(313) type1 = ax.plot(myHist[:,2]) plt.xlabel('iteration') plt.ylabel('X2') plt.show()

import matplotlib import matplotlib.pyplot as plt from numpy import * from matplotlib.patches import Rectangle def loadDataSet(): dataMat = [] labelMat = [] fr = open('F:\machinelearninginaction\Ch05\testSet.txt') for line in fr.readlines(): lineArr = line.strip().split() dataMat.append([1.0, float(lineArr[0]), float(lineArr[1])]) labelMat.append(int(lineArr[2])) return dataMat,labelMat def sigmoid(inX): return 1.0/(1+exp(-inX)) def stocGradAscent0(dataMatrix, classLabels): m,n = shape(dataMatrix) alpha = 0.01 weights = ones(n) #initialize to all ones for i in range(m): h = sigmoid(sum(dataMatrix[i]*weights)) error = classLabels[i] - h weights = weights + alpha * error * dataMatrix[i] return weights def gradAscent(dataMatIn, classLabels): dataMatrix = mat(dataMatIn) #convert to NumPy matrix labelMat = mat(classLabels).transpose() #convert to NumPy matrix m,n = shape(dataMatrix) alpha = 0.001 maxCycles = 500 weights = ones((n,1)) for k in range(maxCycles): #heavy on matrix operations h = sigmoid(dataMatrix*weights) #matrix mult error = (labelMat - h) #vector subtraction weights = weights + alpha * dataMatrix.transpose()* error #matrix mult return weights def stocGradAscent1(dataMatrix, classLabels, numIter=150): m,n = shape(dataMatrix) weights = ones(n) #initialize to all ones for j in range(numIter): dataIndex = range(m) for i in range(m): alpha = 4/(1.0+j+i)+0.0001 #apha decreases with iteration, does not randIndex = int(random.uniform(0,len(dataIndex)))#go to 0 because of the constant h = sigmoid(sum(dataMatrix[randIndex]*weights)) error = classLabels[randIndex] - h weights = weights + alpha * error * dataMatrix[randIndex] # del(dataIndex[randIndex]) return weights dataMat,labelMat=loadDataSet() dataArr = array(dataMat) weights = stocGradAscent1(dataArr,labelMat) n = shape(dataArr)[0] #number of points to create xcord1 = [] ycord1 = [] xcord2 = [] ycord2 = [] markers =[] colors =[] for i in range(n): if int(labelMat[i])== 1: xcord1.append(dataArr[i,1]); ycord1.append(dataArr[i,2]) else: xcord2.append(dataArr[i,1]); ycord2.append(dataArr[i,2]) fig = plt.figure() ax = fig.add_subplot(111) #ax.scatter(xcord,ycord, c=colors, s=markers) type1 = ax.scatter(xcord1, ycord1, s=30, c='red', marker='s') type2 = ax.scatter(xcord2, ycord2, s=30, c='green') x = arange(-3.0, 3.0, 0.1) #weights = [-2.9, 0.72, 1.29] #weights = [-5, 1.09, 1.42] weights = [13.03822793, 1.32877317, -1.96702074] weights = [4.12, 0.48, -0.6168] y = (-weights[0]-weights[1]*x)/weights[2] type3 = ax.plot(x, y) #ax.legend([type1, type2, type3], ["Did Not Like", "Liked in Small Doses", "Liked in Large Doses"], loc=2) #ax.axis([-5000,100000,-2,25]) plt.xlabel('X1') plt.ylabel('X2') plt.show()