
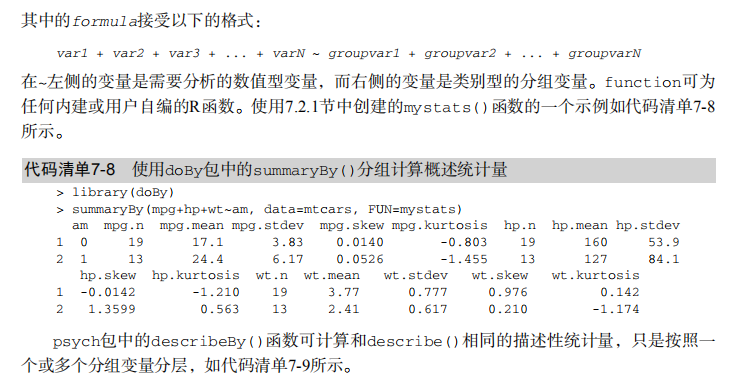
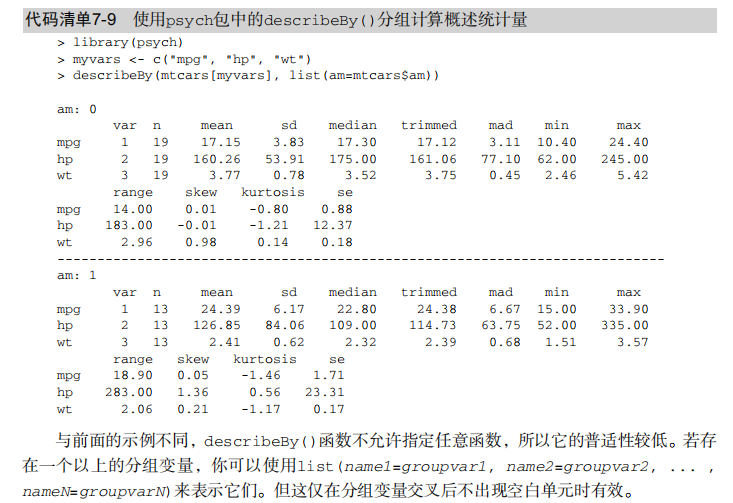

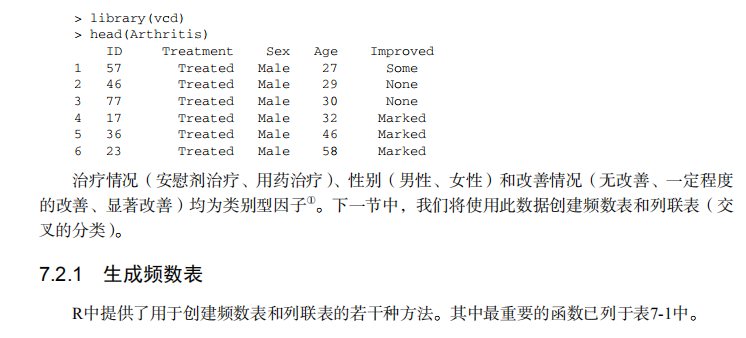




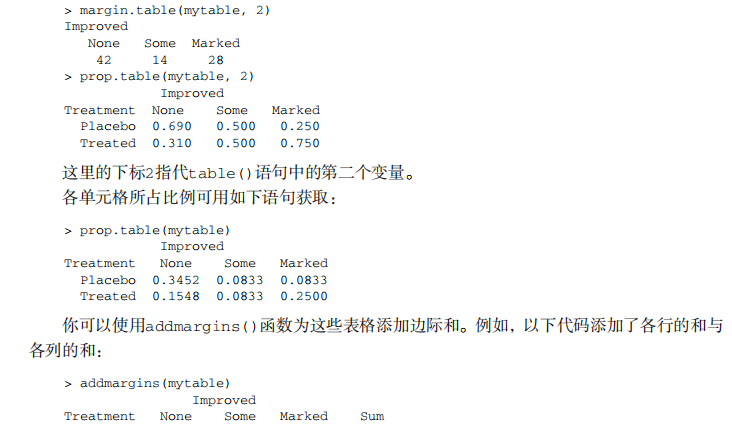
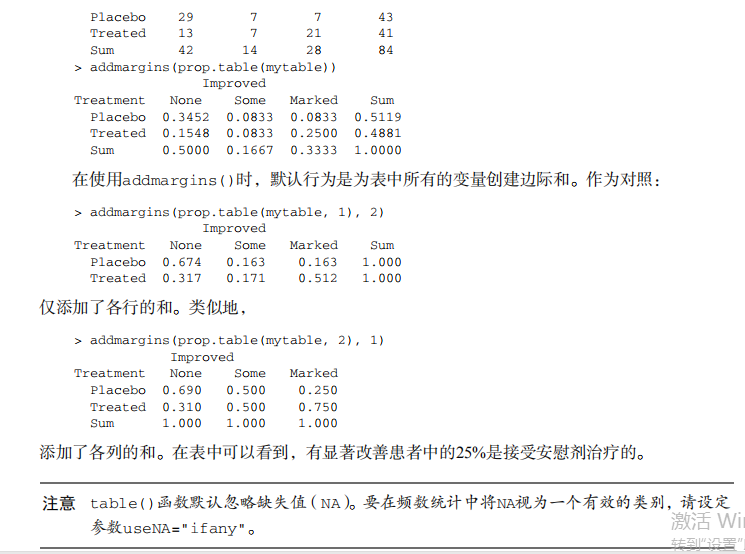
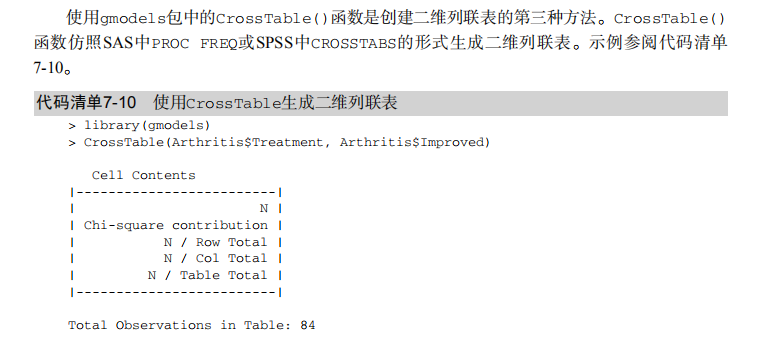
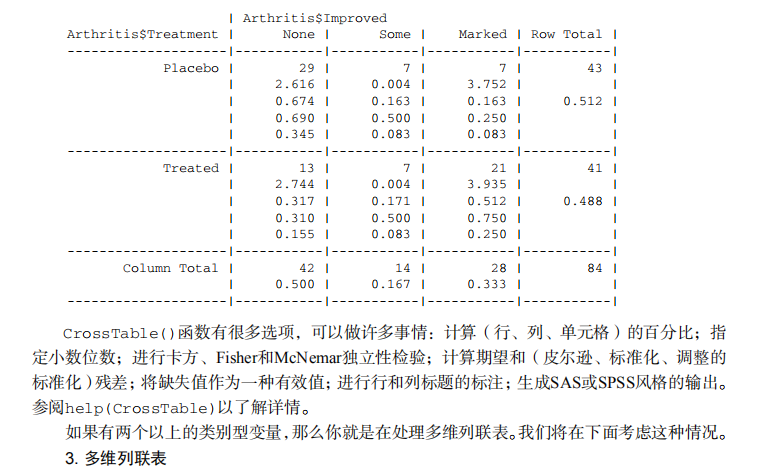
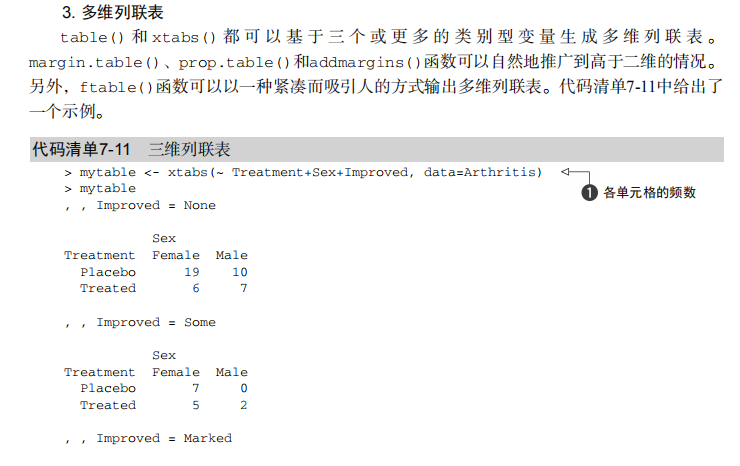
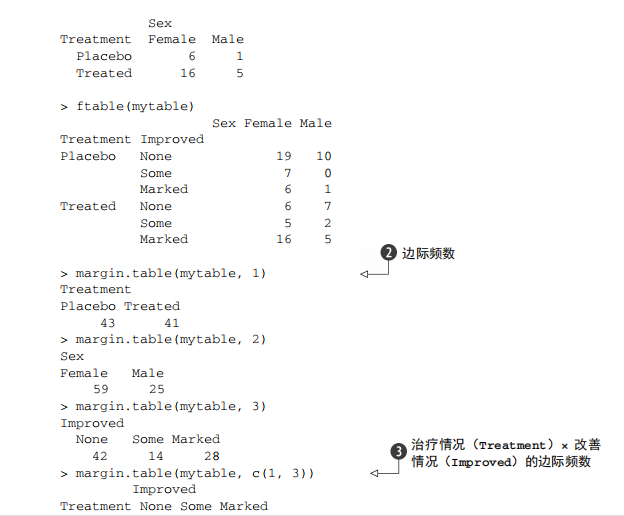
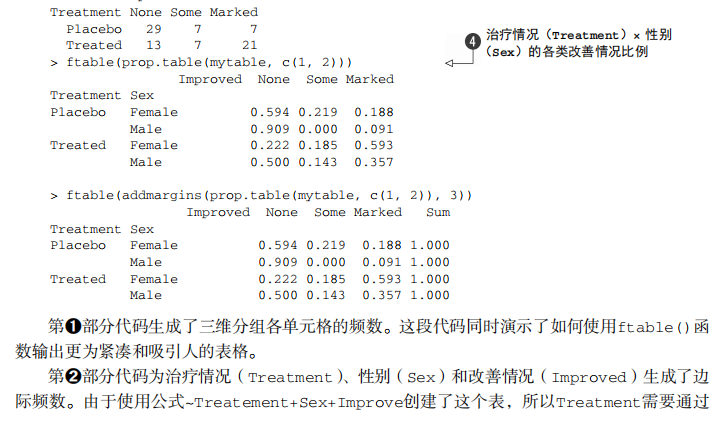
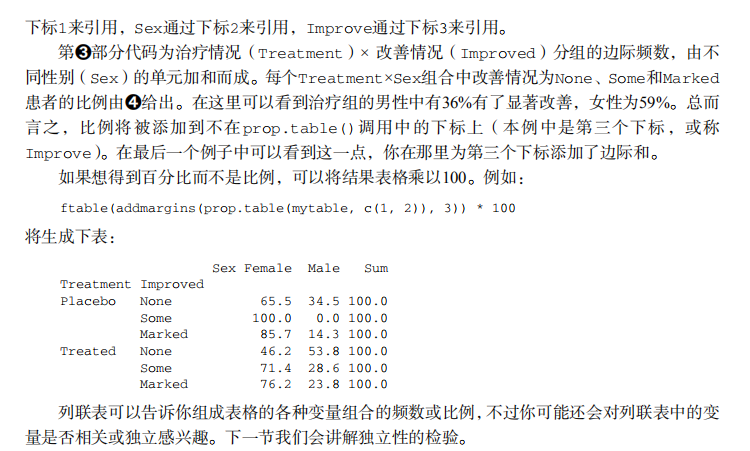
#---------------------------------------------------------------------# # R in Action (2nd ed): Chapter 7 # # Basic statistics # # requires packages npmc, ggm, gmodels, vcd, Hmisc, # # pastecs, psych, doBy to be installed # # install.packages(c("ggm", "gmodels", "vcd", "Hmisc", # # "pastecs", "psych", "doBy")) # #---------------------------------------------------------------------# mt <- mtcars[c("mpg", "hp", "wt", "am")] head(mt) # Listing 7.1 - Descriptive stats via summary mt <- mtcars[c("mpg", "hp", "wt", "am")] summary(mt) # Listing 7.2 - descriptive stats via sapply mystats <- function(x, na.omit=FALSE){ if (na.omit) x <- x[!is.na(x)] m <- mean(x) n <- length(x) s <- sd(x) skew <- sum((x-m)^3/s^3)/n kurt <- sum((x-m)^4/s^4)/n - 3 return(c(n=n, mean=m, stdev=s, skew=skew, kurtosis=kurt)) } myvars <- c("mpg", "hp", "wt") sapply(mtcars[myvars], mystats) # Listing 7.3 - Descriptive stats via describe (Hmisc) library(Hmisc) myvars <- c("mpg", "hp", "wt") describe(mtcars[myvars]) # Listing 7.,4 - Descriptive stats via stat.desc (pastecs) library(pastecs) myvars <- c("mpg", "hp", "wt") stat.desc(mtcars[myvars]) # Listing 7.5 - Descriptive stats via describe (psych) library(psych) myvars <- c("mpg", "hp", "wt") describe(mtcars[myvars]) # Listing 7.6 - Descriptive stats by group with aggregate myvars <- c("mpg", "hp", "wt") aggregate(mtcars[myvars], by=list(am=mtcars$am), mean) aggregate(mtcars[myvars], by=list(am=mtcars$am), sd) # Listing 7.7 - Descriptive stats by group via by dstats <- function(x)sapply(x, mystats) myvars <- c("mpg", "hp", "wt") by(mtcars[myvars], mtcars$am, dstats) # Listing 7.8 - Descriptive stats by group via summaryBy library(doBy) summaryBy(mpg+hp+wt~am, data=mtcars, FUN=mystats) # Listing 7.9 - Descriptive stats by group via describe.by (psych) library(psych) myvars <- c("mpg", "hp", "wt") describeBy(mtcars[myvars], list(am=mtcars$am)) # summary statistics by group via the reshape package library(reshape) dstats <- function(x)(c(n=length(x), mean=mean(x), sd=sd(x))) dfm <- melt(mtcars, measure.vars=c("mpg", "hp", "wt"), id.vars=c("am", "cyl")) cast(dfm, am + cyl + variable ~ ., dstats) # frequency tables library(vcd) head(Arthritis) # one way table mytable <- with(Arthritis, table(Improved)) mytable # frequencies prop.table(mytable) # proportions prop.table(mytable)*100 # percentages # two way table mytable <- xtabs(~ Treatment+Improved, data=Arthritis) mytable # frequencies margin.table(mytable,1) #row sums margin.table(mytable, 2) # column sums prop.table(mytable) # cell proportions prop.table(mytable, 1) # row proportions prop.table(mytable, 2) # column proportions addmargins(mytable) # add row and column sums to table # more complex tables addmargins(prop.table(mytable)) addmargins(prop.table(mytable, 1), 2) addmargins(prop.table(mytable, 2), 1) # Listing 7.10 - Two way table using CrossTable library(gmodels) CrossTable(Arthritis$Treatment, Arthritis$Improved) # Listing 7.11 - Three way table mytable <- xtabs(~ Treatment+Sex+Improved, data=Arthritis) mytable ftable(mytable) margin.table(mytable, 1) margin.table(mytable, 2) margin.table(mytable, 2) margin.table(mytable, c(1,3)) ftable(prop.table(mytable, c(1,2))) ftable(addmargins(prop.table(mytable, c(1, 2)), 3)) # Listing 7.12 - Chi-square test of independence library(vcd) mytable <- xtabs(~Treatment+Improved, data=Arthritis) chisq.test(mytable) mytable <- xtabs(~Improved+Sex, data=Arthritis) chisq.test(mytable) # Fisher's exact test mytable <- xtabs(~Treatment+Improved, data=Arthritis) fisher.test(mytable) # Chochran-Mantel-Haenszel test mytable <- xtabs(~Treatment+Improved+Sex, data=Arthritis) mantelhaen.test(mytable) # Listing 7.13 - Measures of association for a two-way table library(vcd) mytable <- xtabs(~Treatment+Improved, data=Arthritis) assocstats(mytable) # Listing 7.14 Covariances and correlations states<- state.x77[,1:6] cov(states) cor(states) cor(states, method="spearman") x <- states[,c("Population", "Income", "Illiteracy", "HS Grad")] y <- states[,c("Life Exp", "Murder")] cor(x,y) # partial correlations library(ggm) # partial correlation of population and murder rate, controlling # for income, illiteracy rate, and HS graduation rate pcor(c(1,5,2,3,6), cov(states)) # Listing 7.15 - Testing a correlation coefficient for significance cor.test(states[,3], states[,5]) # Listing 7.16 - Correlation matrix and tests of significance via corr.test library(psych) corr.test(states, use="complete") # t test library(MASS) t.test(Prob ~ So, data=UScrime) # dependent t test sapply(UScrime[c("U1","U2")], function(x)(c(mean=mean(x),sd=sd(x)))) with(UScrime, t.test(U1, U2, paired=TRUE)) # Wilcoxon two group comparison with(UScrime, by(Prob, So, median)) wilcox.test(Prob ~ So, data=UScrime) sapply(UScrime[c("U1", "U2")], median) with(UScrime, wilcox.test(U1, U2, paired=TRUE)) # Kruskal Wallis test states <- data.frame(state.region, state.x77) kruskal.test(Illiteracy ~ state.region, data=states) # Listing 7.17 - Nonparametric multiple comparisons source("http://www.statmethods.net/RiA/wmc.txt") states <- data.frame(state.region, state.x77) wmc(Illiteracy ~ state.region, data=states, method="holm")