在Ubuntu上使用libsvm(附上官网链接以及安装方法)进行SVM的实践:
1、代码演示:(来自一段文本分类的代码)
# encoding=utf8
__author__ = 'wang'
# set the encoding of input file utf-8
import sys
reload(sys)
sys.setdefaultencoding('utf-8')
import os
from svmutil import *
import subprocess
# get the data in svm format
featureStringCorpusDir = './svmFormatFeatureStringCorpus/'
fileList = os.listdir(featureStringCorpusDir)
# get the number of the files
numFile = len(fileList)
trainX = []
trainY = []
testX = []
testY = []
# enumrate the file
for i in xrange(numFile):
# print i
fileName = fileList[i]
if(fileName.endswith(".libsvm")):
# check the data format
#
checkdata_py = r"./tool/checkdata.py"
# the file name
svm_file = featureStringCorpusDir + fileName
cmd = 'python {0} {1} '.format(checkdata_py, svm_file)
# print ("excute '{}'".format(cmd))
# print('Check Data...')
check_data = subprocess.Popen(cmd, shell = True, stdout = subprocess.PIPE)
(stdout, stderr) = check_data.communicate()
returncode = check_data.returncode
if returncode == 1:
print stdout
exit(1)
y, x = svm_read_problem(svm_file)
trainX += x[:100]
trainY += y[:100]
testX += x[-1:-6:-1]
testY += y[-1:-6:-1]
print testX
# train the c-svm model
m = svm_train(trainY, trainX, '-s 0 -c 4 -t 2 -g 0.1 -e 0.1')
# predict the test data
p_label, p_acc, p_val = svm_predict(testY, testX, m)
# output the result to the file
result_file = open("./result.txt","w")
result_file.write( "testY p_label p_val
")
for c in xrange(len(p_label)):
result_file.write( str(testY[c]) + " " + str(p_label[c]) + " " + str(p_val[c]) + "
")
result_file.write( "accuracy: " + str(p_acc) + "
")
result_file.close()
# print p_val
(1)关于训练数据的格式:
The format of training and testing data file is: <label> <index1>:<value1> <index2>:<value2> .. . . . Each line contains an instance and is ended by a ' ' character. For
classification, <label> is an integer indicating the class label (multi-class is supported)(如何label是个非数值的,将其转化为数值型). For regression, <label> is
the target value which can be any real number. For one-class SVM, it's not used so can be any number. The pair <index>:<value> gives a feature (attribute) value:
<index> is an integer starting from 1 and <value> is a real number. The only exception
is the precomputed kernel, where <index> starts from 0; see the section of precomputed kernels.
Indices must be in ASCENDING order(要升序排序). Labels in the testing file are only used to calculate accuracy
or errors. If they are unknown, just fill the first column with any numbers. A sample classification data included in this package is `heart_scale'. To check if your data is
in a correct form, use `tools/checkdata.py' (details in `tools/README'). Type `svm-train heart_scale', and the program will read the training data and output the model file `heart_scale.model'. If you have a test set called heart_scale.t, then type `svm-predict heart_scale.t heart_scale.model output' to see the prediction accuracy. The `output' file contains the predicted class labels. For classification, if training data are in only one class (i.e., all labels are the same), then `svm-train' issues a warning message: `Warning: training data in only one class. See README for details,' which means the training data is very unbalanced. The label in the training data is directly returned when testing.
(2)svm训练的参数解释来自官方的github:https://github.com/cjlin1/libsvm
`svm-train' Usage
=================
Usage: svm-train [options] training_set_file [model_file]
options:
-s svm_type : set type of SVM (default 0)
0 -- C-SVC (multi-class classification)
1 -- nu-SVC (multi-class classification)
2 -- one-class SVM
3 -- epsilon-SVR (regression)
4 -- nu-SVR (regression)
-t kernel_type : set type of kernel function (default 2)
0 -- linear: u'*v
1 -- polynomial: (gamma*u'*v + coef0)^degree
2 -- radial basis function: exp(-gamma*|u-v|^2)
3 -- sigmoid: tanh(gamma*u'*v + coef0)
4 -- precomputed kernel (kernel values in training_set_file)
-d degree : set degree in kernel function (default 3)
-g gamma : set gamma in kernel function (default 1/num_features)
-r coef0 : set coef0 in kernel function (default 0)
-c cost : set the parameter C of C-SVC, epsilon-SVR, and nu-SVR (default 1)
-n nu : set the parameter nu of nu-SVC, one-class SVM, and nu-SVR (default 0.5)
-p epsilon : set the epsilon in loss function of epsilon-SVR (default 0.1)
-m cachesize : set cache memory size in MB (default 100)
-e epsilon : set tolerance of termination criterion (default 0.001)
-h shrinking : whether to use the shrinking heuristics, 0 or 1 (default 1)
-b probability_estimates : whether to train a SVC or SVR model for probability estimates, 0 or 1 (default 0)
-wi weight : set the parameter C of class i to weight*C, for C-SVC (default 1) (当数据不平衡时,可给不同的类设置不用的惩罚)
-v n: n-fold cross validation mode
-q : quiet mode (no outputs)
The k in the -g option means the number of attributes in the input data.
option -v randomly splits the data into n parts and calculates cross
validation accuracy/mean squared error on them.
See libsvm FAQ for the meaning of outputs.
`svm-predict' Usage
===================
Usage: svm-predict [options] test_file model_file output_file
options:
-b probability_estimates: whether to predict probability estimates, 0 or 1 (default 0); for one-class SVM only 0 is supported
model_file is the model file generated by svm-train.
test_file is the test data you want to predict.
svm-predict will produce output in the output_file.
注:
nu-SVR: nu回归机。引入能够自动计算epsilon的参数nu。若记错误样本的个数为q ,则nu大于等于q/l,即nu是错误样本的个数所占总样本数的份额的上界;若记支持向量的个数为p,则nu小于等于p/l,即nu是支持向量的个数所占总样本数的份额的下界。首先选择参数nu和C,然后求解最优化问题。
Shrinking: 即参数-h, 优化求解过程中是否采用shrinking. 边界支持向量BSVs(ai=C的SV)在迭代过程中ai不会变化,如果找到这些点,并把它们固定为C,可以减少QP的规模。
2、运行结果展示并分析部分参数的意思:
* optimization finished, #iter = 119 nu = 0.272950 obj = -122.758206, rho = -1.384603 nSV = 127, nBSV = 21 * ..............(此处省略部分) * optimization finished, #iter = 129 nu = 0.214275 obj = -89.691907, rho = -1.105131 nSV = 103, nBSV = 8 * optimization finished, #iter = 77 nu = 0.147922 obj = -67.825431, rho = 0.984237 nSV = 80, nBSV = 10 Total nSV = 1246 Accuracy = 51.4286% (36/70) (classification)
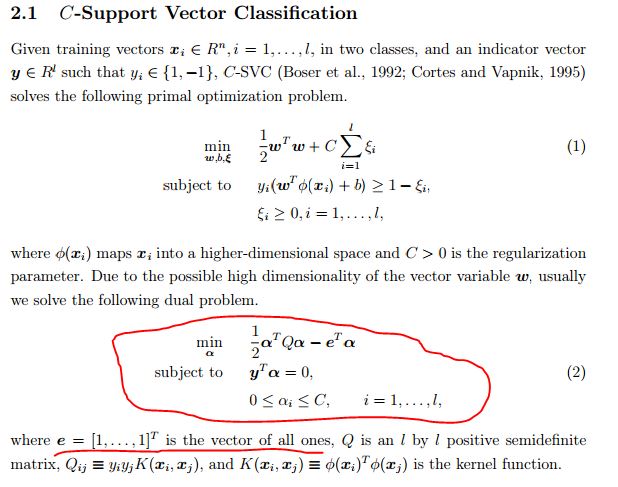
官方解释:obj is the optimal objective value of the dual SVM problem(obj表示SVM最优化问题对偶式的最优目标值,公式如下图的公式(2)). rho is the bias term in the decision function sgn(w^Tx - rho). nSV and nBSV are number of support vectors and bounded support vectors (i.e., alpha_i = C). nu-svm is a somewhat equivalent form of C-SVM where C is replaced by nu. nu simply shows the corresponding parameter. More details are in libsvm document.