一.聚类:
一般步骤:
1.选择合适的变量
2.缩放数据
3.寻找异常点
4.计算距离
5.选择聚类算法
6.采用一种或多种聚类方法
7.确定类的数目
8.获得最终聚类的解决方案
9.结果可视化
10.解读类
11.验证结果
1.层次聚类分析
案例:采用flexclust的营养数据集作为参考
1.基于5种营养标准的27类鱼,禽,肉的相同点和不同点是什么
2.是否有一种办法把这些食物分成若干各类
1.1计算距离
1 data(nutrient,package = 'flexclust') 2 head(nutrient,4) 3 d <- dist(nutrient) 4 as.matrix(d)[1:4,1:4]
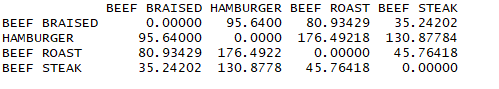
结论:观测的距离越大,异质性越大
1.2平均联动聚类
1 row.names(nutrient) <- tolower(row.names(nutrient)) 2 nutrient.scaled <- scale(nutrient) 3 d2 <- dist(nutrient.scaled) 4 fit.average <- hclust(d2,method = 'average') 5 plot(fit.average,hang=-1,cex=.8,main='Average Linkage Clustering')
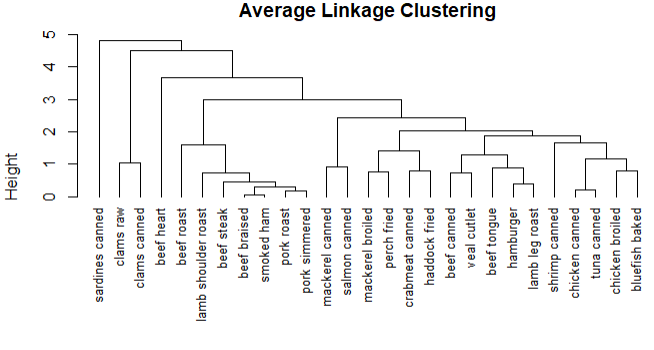
结论:只能提供食物营养成分的相似性和相异性
1.3获取聚类的个数
1 library('NbClust') 2 devAskNewPage(ask = T) 3 nc <- NbClust(nutrient.scaled, distance="euclidean", 4 min.nc=2, max.nc=15, method="average") 5 table(nc$Best.n[1,]) 6 barplot(table(nc$Best.n[1,]), 7 xlab = 'Number of Clusters',ylab = 'Number of Criteria', 8 main='Number of Clusters chosen by 26 criteria')

结论:分别有4个投票数最多的聚类(2,3,5,15),从中选择一个更适合的聚类数
1.4获取聚类的最终方案
1 # 聚类分配情况 2 clusters <- cutree(fit.average,k=5) 3 table(clusters) 4 # 描述聚类 5 aggregate(nutrient,by=list(clusters=clusters),median) 6 aggregate(as.data.frame(nutrient.scaled),by=list(clusters=clusters),median) 7 plot(fit.average,hang=-1,cex=.8,main='Average Linkpage Clustering 5 Cluster Solution') 8 rect.hclust(fit.average,k=5)
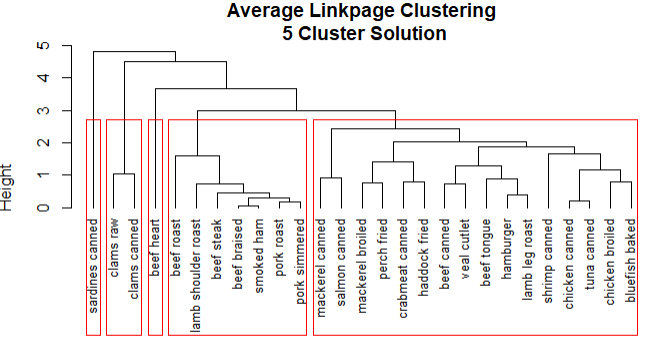
结论:
1.sardines canned形成自己的类,因为钙含量比较高
2.beef heart也是单独的类,富含蛋白质和铁
3.beef roast到pork simmered含有高能量和脂肪
4.clams raw到clams canned含有较高的维生素
5.mackerel canned到bluefish baked含有较低的铁
2.划分聚类分析
案例:采用rattle.data中的wine数据集进行分析
1.葡萄酒数据的K均值聚类
# 使用卵石图确定类的数量 wssplot <- function(data,nc=15,seed=1234){ wss <- (nrow(data)-1) * sum(apply(data,2,var)) for (i in 2:nc) { set.seed(seed) wss[i] <- sum(kmeans(data,centers = i)$withinss) } plot(1:nc,wss,type = 'b',xlab = 'Number of Clusters',ylab = 'Within groups sum of squares') }
1 data(wine,package = 'rattle.data') 2 3 head(wine) 4 df <- scale(wine[-1]) 5 wssplot(df) 6 library(NbClust) 7 set.seed(1234) 8 # 确定聚类的数量 9 nc <- NbClust(df, min.nc=2, max.nc=15, method="kmeans") 10 table(nc$Best.n[1,]) 11 barplot(table(nc$Best.n[1,]), 12 xlab="Numer of Clusters", ylab="Number of Criteria", 13 main="Number of Clusters Chosen by 26 Criteria") 14 set.seed(1234) 15 # 进行k值聚类分析 16 fit.km <- kmeans(df, 3, nstart=25) 17 fit.km$size 18 fit.km$centers 19 aggregate(wine[-1], by=list(cluster=fit.km$cluster), mean)

结论:分3个聚类对数据有很好的拟合
# 使用兰德系数来量化类型变量和类之间的协议 ct.km <- table(wine$Type,fit.km$cluster) library(flexclust) randIndex(ct.km)

结论:拟合结果优秀
围绕中心点的分类:因为K均值聚类方法是基于均值的,所以对异常值较为敏感,更为稳健的方法是围绕中心点的划分,
k均值聚类一般使用欧几里得距离,而PAM可以使用任意的距离来计算
1 library(cluster) 2 set.seed(1234) 3 fit.pam <- pam(wine[-1],k=3,stand = T) 4 fit.pam$medoids 5 clusplot(fit.pam,main = 'Bivariate Cluster Plot') 6 ct.pam <- table(wine$Type,fit.pam$clustering) 7 randIndex(ct.pam)
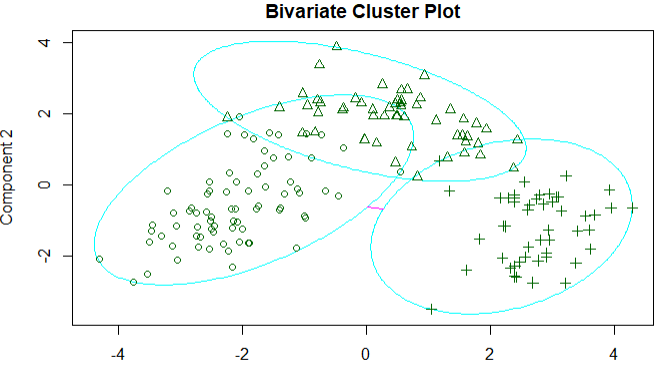
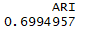
结论:调整后的兰德指数从之前的0.9下降到0.7
3.避免不存在的聚类
3.1查看数据集
1 library(fMultivar) 2 set.seed(1234) 3 df <- rnorm2d(1000,rho=.5) 4 df <- as.data.frame(df) 5 plot(df,main='Bivariate Normal Distribution with rho=0.5')

结论:没有存在的类
3.2计算聚类的个数
library(NbClust) nc <- NbClust(df,min.nc = 2,max.nc = 15,method = 'kmeans') dev.new() barplot(table(nc$Best.n[1,]),xlab="Numer of Clusters", ylab="Number of Criteria", main="Number of Clusters Chosen by 26 Criteria")

结论:一共可分为3各类
3.3聚类图像
1 library(ggplot2) 2 fit2 <- pam(df,k=2) 3 df$clustering <- factor(fit2$clustering) 4 ggplot(data = df,aes(x=V1,y=V2,color=clustering,shape=clustering))+ 5 geom_point()+ 6 ggtitle('Clustering of Bivariate Normal Data')
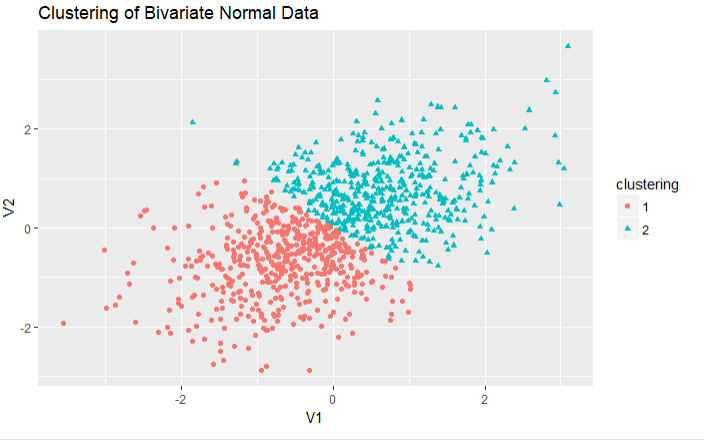
结论:对于二元数据的PAM聚类分析,提取出2类
3.4分析聚类
plot(nc$All.index[,4],type='o',ylab='ccc',xlab='Number of Clusters',col='blue')
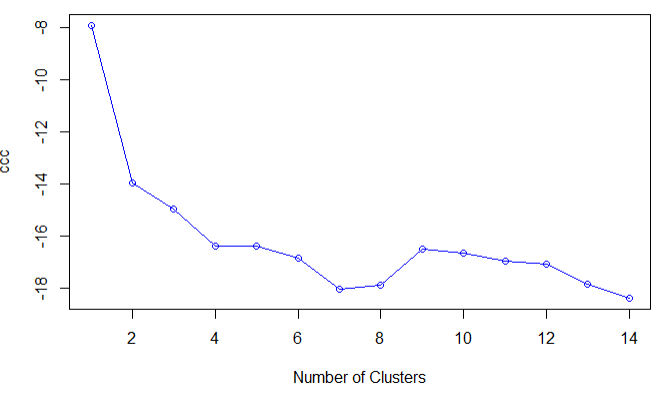
结论:二元正态数据的CCC图,表明没有类存在,当CCC为负数并且对于两类或者是更多的类的递减
二.分类
使用机器学习来预测二分类结果
案例分析:使用乳腺癌数据作为测试,训练集建立逻辑回归,决策时,条件推断树,随机森林,支持向量机等分类模型,测试集用于评估各个模型的有效性
1.准备数据
loc <- "http://archive.ics.uci.edu/ml/machine-learning-databases/" ds <- "breast-cancer-wisconsin/breast-cancer-wisconsin.data" url <- paste(loc, ds, sep="") breast <- read.table(url, sep=",", header=FALSE, na.strings="?") names(breast) <- c("ID", "clumpThickness", "sizeUniformity", "shapeUniformity", "maginalAdhesion", "singleEpithelialCellSize", "bareNuclei", "blandChromatin", "normalNucleoli", "mitosis", "class") df <- breast[-1] df$class <-factor(df$class,levels = c(2,4),labels = c('begign','malignant')) set.seed(1234) train <- sample(nrow(df),0.7*nrow(df)) df.train <- df[train,] df.validate <- df[-train,] table(df.train$class) table(df.validate$class)
2.逻辑回归
# 拟合逻辑回归 fit.logit <- glm(class~.,data=df.train,family = binomial()) prob <- predict(fit.logit,df.validate,type='response') # 对训练集外的样本进行分类 logit.pred <- factor(prob>.5,levels = c(F,T),labels = c('benign','malignant')) # 评估预测的准确性 logit.pref <- table(df.validate$class,logit.pred,dnn = c('Actual','Predicted')) logit.pref

结论:正确分类的模型是97%
3.决策树
library(rpart) set.seed(1234) # 生成树 dtree <- rpart(class~.,data = df.train,method = 'class',parms = list(split='information')) plotcp(dtree) # 剪枝 dtree.pruned <- prune(dtree,cp=.0125) library(rpart.plot) prp(dtree.pruned,type = 2,extra = 104,fallen.leaves = T,main='decision tree') # 对训练集外的样本单元分类 dtree.pred <- predict(dtree.pruned,df.validate,type='class') dtree.pref <- table(df.validate$class,dtree.pred,dnn = c('Actual','Predict')) dtree.pref
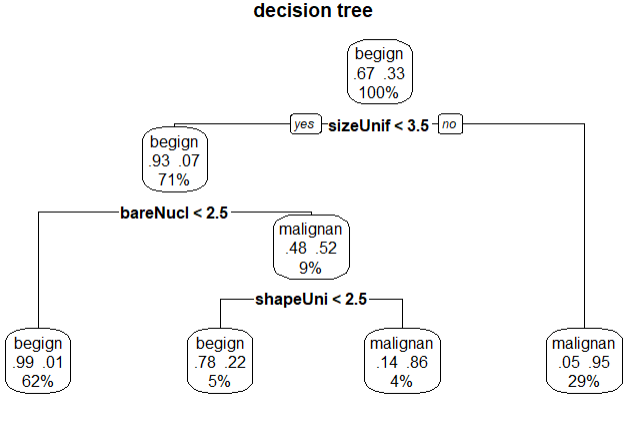
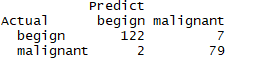
结论:验证的准确率96%
4.条件推断树
library(party) library(partykit) fit.tree <- ctree(class~.,data=df.train) plot(fit.tree,main='Condition Inference Tree') ctree.pred <- predict(fit.tree,df.validate,type='response') ctree.pref <- table(df.validate$class,ctree.pred,dnn = c('Actual','Predicted')) ctree.pref
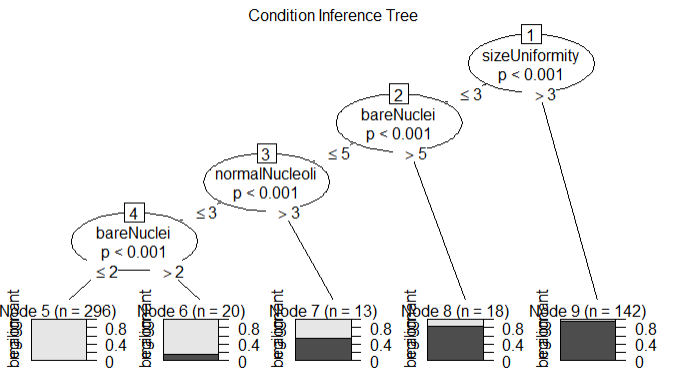
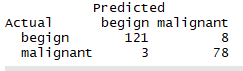
结论:验证的准确率97%
5.随机森林
library(randomForest) set.seed(1234) # 生成森林 fit.forest <- randomForest(class~.,data=df.train,na.action=na.roughfix,importance=T) importance(fit.forest,type=2) forest.pred <- predict(fit.forest,df.validate) # 对训练集外的样本点分类 forest.pref <- table(df.validate$class,forest.pred,dnn = c('Actual','Predicted')) forest.pref

结论:验证准确率在98%
6.支持向量机
library(e1071) set.seed(1234) fit.svm <- svm(class~.,data=df.train) svm.pred <- predict(fit.svm,na.omit(df.validate)) svm.pref <- table(na.omit(df.validate)$class,svm.pred,dnn = c('Actual','Predicted')) svm.pref

结论:验证准确率在96%
7.带有RBF内核的支持向量机
set.seed(1234) # 通过调整gamma和c来拟合模型 tuned <- tune.svm(class~.,data=df.train,gamma = 10^(-6:1),cost = 10^(-10:10)) tuned fit.svm <- svm(class~.,data=df.train,gamma=.01,cost=1) svm.pred <- predict(fit.svm,na.omit(df.validate)) svm.pref <- table(na.omit(df.validate)$class,svm.pred,dnn = c('Actual','Predicted')) svm.pref

结论验证的成功率有97%
8.编写函数选择预测效果最好的解
performance <- function(table,n=2){ if(!all(dim(table) == c(2,2))){ stop('Must be a 2 * 2 table') } tn = table[1,1] fp = table[1,2] fn = table[2,1] tp = table[2,2] sensitivity = tp/(tp+fn) specificity = tn/(tn+fp) ppp = tp/(tp+fp) npp = tn/(tn+fn) hitrate = (tp+tn)/(tp+tn+fn+fp) result <- paste("Sensitivity = ", round(sensitivity, n) , " Specificity = ", round(specificity, n), " Positive Predictive Value = ", round(ppp, n), " Negative Predictive Value = ", round(npp, n), " Accuracy = ", round(hitrate, n), " ", sep="") cat(result) }
performance(logit.pref)
performance(dtree.pref)
performance(ctree.pref)
performance(forest.pref)
performance(svm.pref)

结论:从以上的分类器中,本案例随机森林的拟合度最优
三.使用rattle进行数据挖掘
案例:预测糖尿病
library(rattle)
rattle()

结论:设定好这些变量点击执行

选择model选项卡,然后选择条件推断树作为预测模型,点击Draw生成图片

通过Evalute选项卡来评估模型

结论:只有35%的病人被成功鉴别,我们可以试试随机森林和支持向量机的匹配度是否更高