第一次听到这个概念,很懵,只知道是FLT3基因的内部段串联重复,因为之前是做遗传病相关,也接触过段串联重复序列,
后来经过文献阅读,发现完全不是一回事情。
遗传病中的短串联重复序列:(摘自百度百科)
短串联重复序列 (STR, short tandem repeats) STR: 短串联重复序列(short tandem repeats,STR)也称微卫星 DNA(microsatellite DNA),
通常是基因组中由1~6个碱基单元组成的一段DNA重复序列,由于核心单位重复数目在个体间呈高度变异性并且数量丰富,构成了STR基因座的遗传多态性。
一般认为人类基因组平均每15 kb就存在一个STR基因座。人类基因组DNA中平均每6~10kb就有一个STR位点,
其多态性成为法医物证检验个人识别和亲子鉴定的丰富来源。
不同人体基因组卫星DNA重复单位的数目是可变的,因此,形成了极其复杂的等位基因片段长度多态性。
白血病中FLT3/ITD串联重复序列:
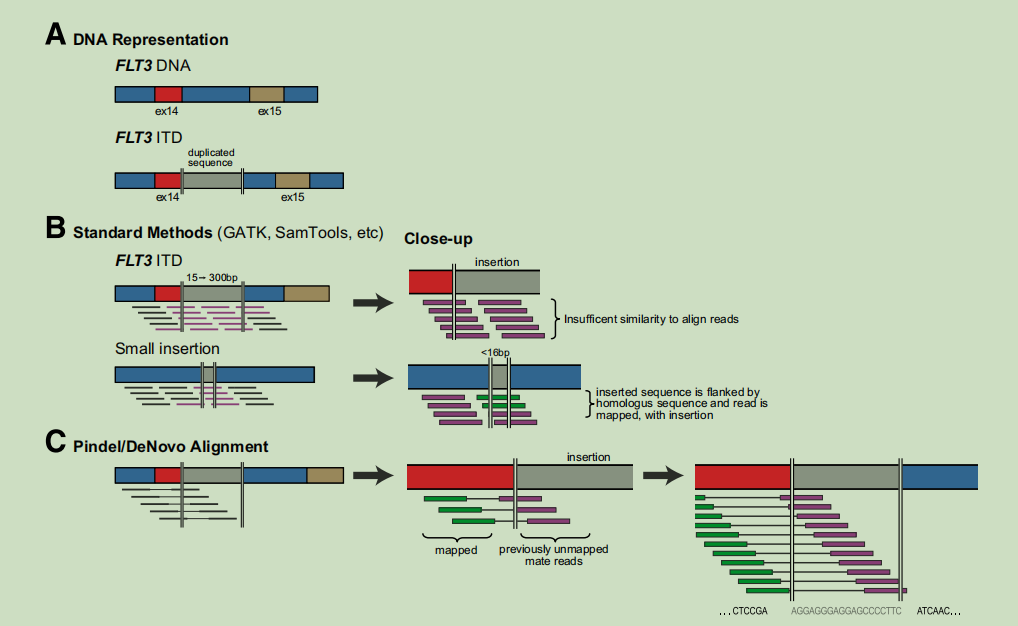
Informatics of insertion detection. A: The FLT3 ITD is an insertion of duplicated and nonduplicated sequence that occurs between exons 13 and
14. These insertions (shown in gray) range in size from 15 to w300 bp. B: Standard methods for fifinding insertions, including the Genome Analysis Toolkit
(GATK), SAMtools, and Dindel, apply probabilistic models to make insertion calls based on data obtained during the initial read mapping and alignment
process. Because of the diffificulty associated with aligning short reads, only small insertion events (generally <15% of the total read length) can be identifified
by this approach (aligned reads are shown in green; unaligned reads, in purple). Such reads generally have suffificient homology in the regions flflanking the
insertion to permit accurate alignment. Large insertions (>16 bp), including the FLT3 ITD, are too long to be detected by this method. C: Using a paired-end
approach, software such as Pindel and de novo alignment can reliably detect larger insertions, including the FLT3 ITD. In this approach, mate-pairs are
identifified in which one end is mapped, but the other is not. The unmapped mates are then assembled to form contigs with partial homology to the reference
sequence, using a pattern-growth algorithm (Pindel) or de novo assembly with a custom script (unpublished data) executing Phrap assembly software. This
method allows for the detection of much larger insertions.
参考文献:
Spencer D H , Abel H J , Lockwood C M , et al. Detection of FLT3 Internal Tandem Duplication in Targeted, Short-Read-Length, Next-Generation Sequencing Data[J]. The Journal of molecular diagnostics: JMD, 2012, 15(1).
