All DNA is composed of a series of nucleotides abbreviated as A, C, G, and T, for example: "ACGAATTCCG". When studying DNA, it is sometimes useful to identify repeated sequences within the DNA.
Write a function to find all the 10-letter-long sequences (substrings) that occur more than once in a DNA molecule.
Example:
Input: s = "AAAAACCCCCAAAAACCCCCCAAAAAGGGTTT"
Output: ["AAAAACCCCC", "CCCCCAAAAA"]
题意:
DNA序列里有ATCG四种,求所有长度为10,出现次数超过一次的序列。
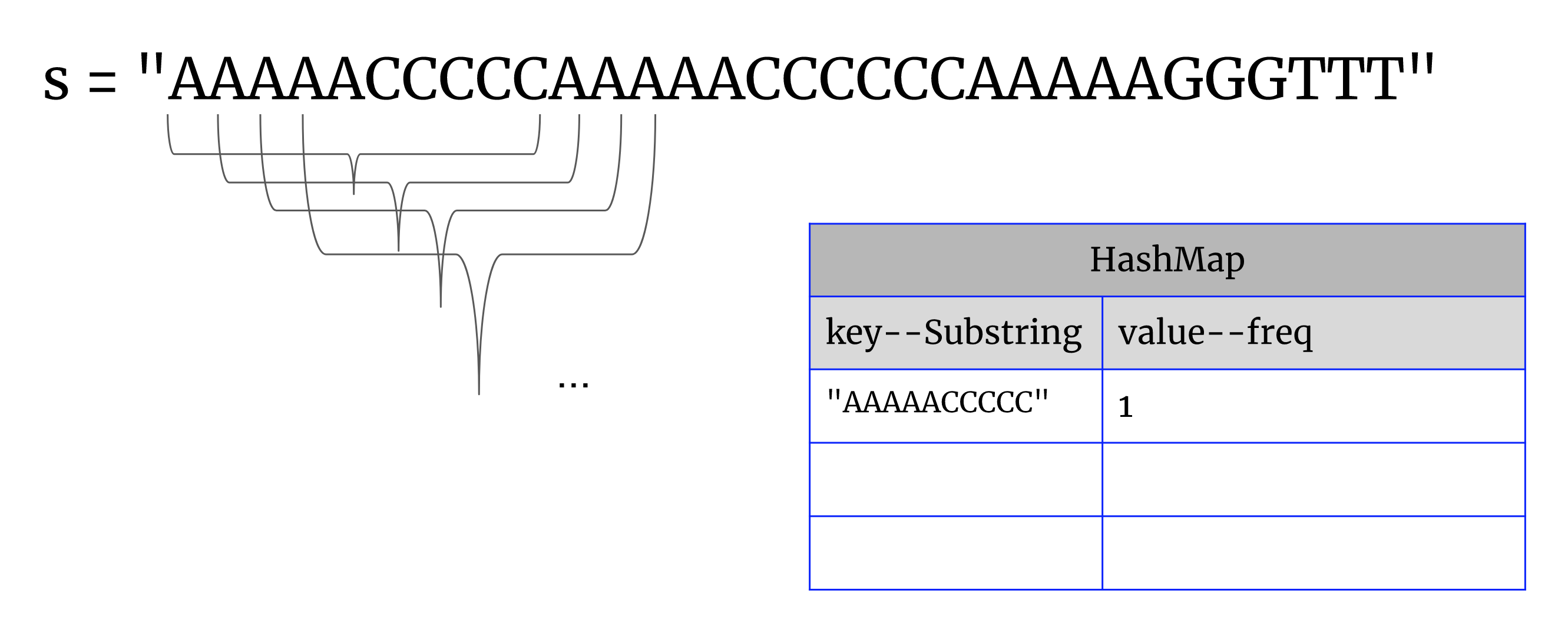
Solution1: HashMap
1. scan the given string, put each 10-letter-long substring and its corresponding frequency into a map
2. looping each entrySet in the map, find if entry.getValue() > 1
code
1 /* 2 Time Complexity: O(n) 3 Space Complexity: O(n) 4 */ 5 class Solution { 6 public List<String> findRepeatedDnaSequences(String s) { 7 List<String> result = new ArrayList<>(); 8 // corner case 9 if (s.length() < 10) return result; 10 11 Map<String, Integer> map = new HashMap<>(); 12 for (int i = 0; i < s.length() - 9; ++i) { 13 String key = s.substring(i, i + 10); 14 if(map.containsKey(key)){ 15 map.put(key, map.get(key) + 1); 16 }else{ 17 map.put(key, 1); 18 } 19 } 20 21 for (Map.Entry<String, Integer> entry : map.entrySet()) { 22 if (entry.getValue() > 1) { 23 result.add(entry.getKey()); 24 } 25 } 26 return result; 27 } 28 }