001、plink
root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped root@PC1:/home/test# plink --file gwas_case_cont --logistic dominant beta 1> /dev/null ## 使用显性模型分析 root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink.assoc.logistic plink.log root@PC1:/home/test# head -n 5 plink.assoc.logistic CHR SNP BP A1 TEST NMISS BETA STAT P 1 snp1 3046 A DOM 288 0.1018 0.2601 0.7948 1 snp2 3092 T DOM 288 0.1018 0.2601 0.7948 1 snp3 3174 T DOM 288 -0.04925 -0.159 0.8737 1 snp4 32399 T DOM 288 0.0702 0.2153 0.8295
002、R语言
root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped root@PC1:/home/test# plink --file gwas_case_cont --recode A 1> /dev/null ## 转换为数值型 root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink.log plink.raw root@PC1:/home/test# sed 1d plink.raw | cut -d " " -f 7- | sed 's/2/1/g' > temp root@PC1:/home/test# sed -n 1p plink.raw | cat - <(sed 1d plink.raw | cut -d " " -f 1-6 | paste -d " " - temp ) > plink2.raw ## 修改格式 root@PC1:/home/test# ls gwas_case_cont.map gwas_case_cont.ped plink2.raw plink.log plink.raw temp root@PC1:/home/test# head plink2.raw | cut -d " " -f 1-10 FID IID PAT MAT SEX PHENOTYPE snp1_A snp2_T snp3_T snp4_T A1 A1 0 0 1 1 1 1 0 0 A2 A2 0 0 1 1 0 0 0 0 A3 A3 0 0 1 1 0 0 0 0 A4 A4 0 0 1 1 0 0 0 0 A5 A5 0 0 1 1 0 0 0 0 A6 A6 0 0 1 1 0 0 0 0 A7 A7 0 0 1 1 0 0 0 0 A8 A8 0 0 1 1 0 0 0 0 A9 A9 0 0 1 1 0 0 0 0
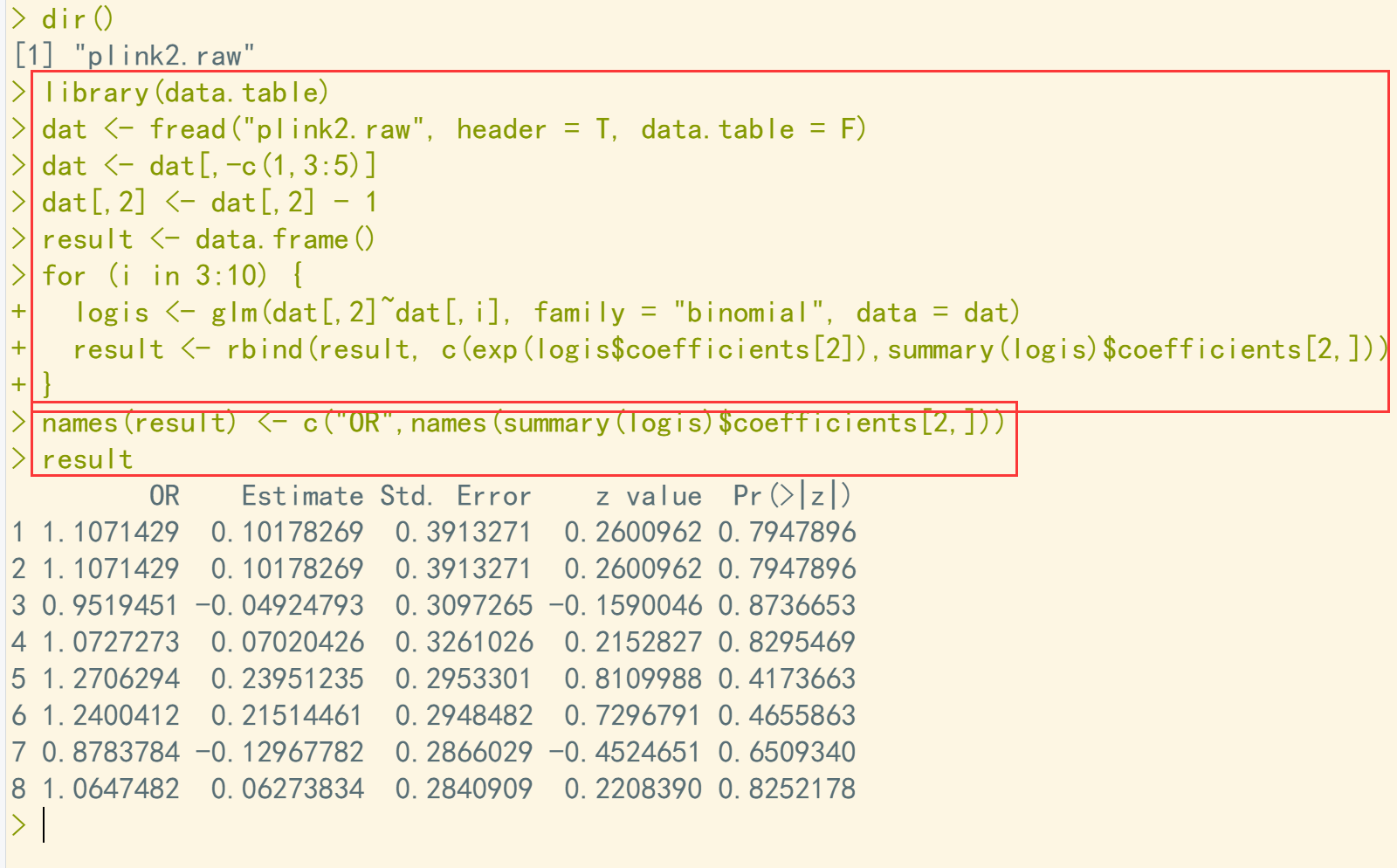
dir() library(data.table) dat <- fread("plink2.raw", header = T, data.table = F) dat <- dat[,-c(1,3:5)] dat[,2] <- dat[,2] - 1 result <- data.frame() for (i in 3:10) { logis <- glm(dat[,2]~dat[,i], family = "binomial", data = dat) result <- rbind(result, c(exp(logis$coefficients[2]),summary(logis)$coefficients[2,])) } names(result) <- c("OR",names(summary(logis)$coefficients[2,])) result