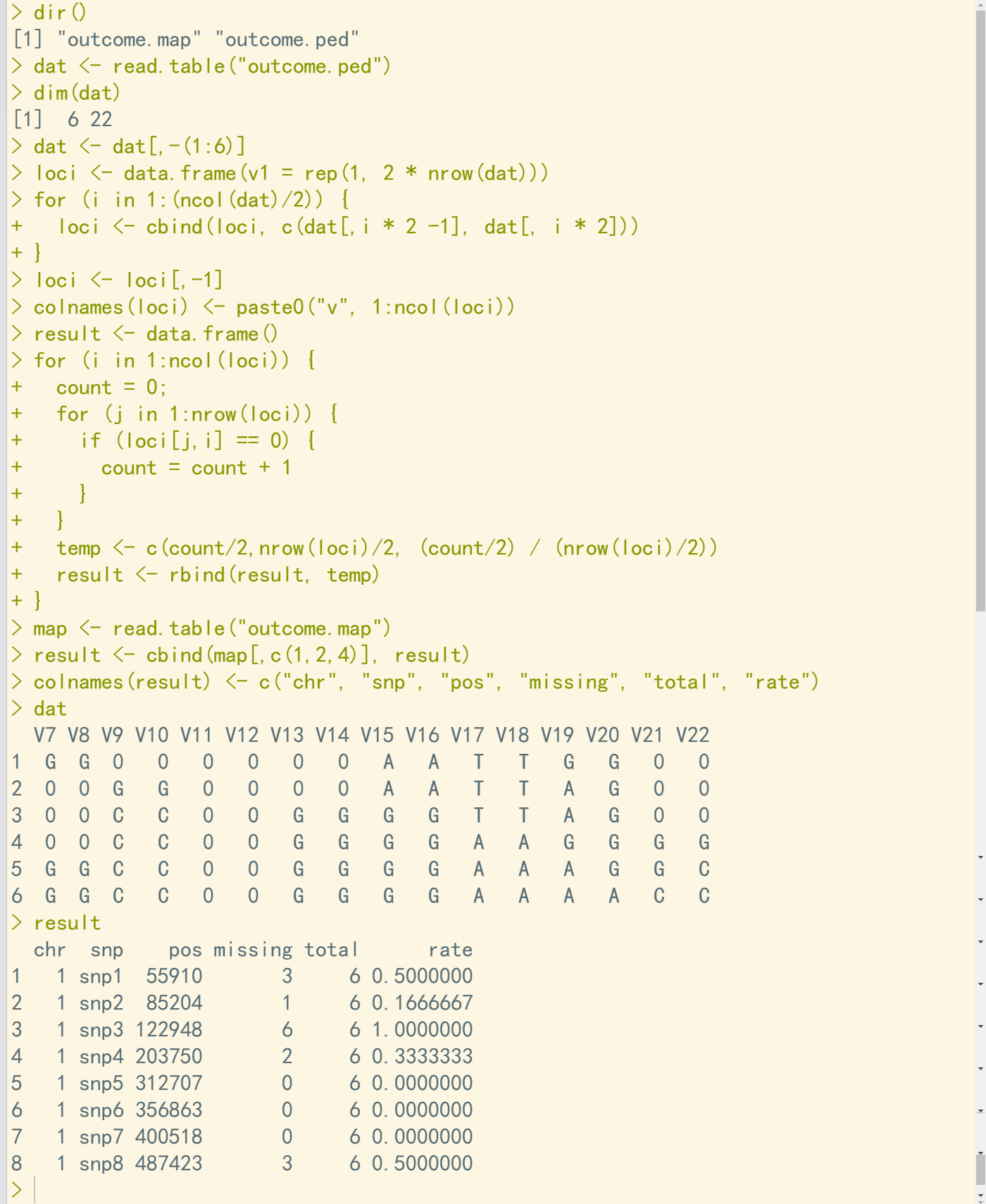
1、R实现
dir() dat <- read.table("outcome.ped") dim(dat) dat <- dat[,-(1:6)] loci <- data.frame(v1 = rep(1, 2 * nrow(dat))) for (i in 1:(ncol(dat)/2)) { loci <- cbind(loci, c(dat[,i * 2 -1], dat[, i * 2])) } loci <- loci[,-1] colnames(loci) <- paste0("v", 1:ncol(loci)) result <- data.frame() for (i in 1:ncol(loci)) { count = 0; for (j in 1:nrow(loci)) { if (loci[j,i] == 0) { count = count + 1 } } temp <- c(count/2,nrow(loci)/2, (count/2) / (nrow(loci)/2)) result <- rbind(result, temp) } map <- read.table("outcome.map") result <- cbind(map[,c(1,2,4)], result) colnames(result) <- c("chr", "snp", "pos", "missing", "total", "rate") dat result
2、plink软件验证
[root@centos79 test]# plink --file outcome --missing --out temp > /dev/null; rm *.log *.nosex [root@centos79 test]# ls missresult.txt outcome.map outcome.ped temp.imiss temp.lmiss test test.sh [root@centos79 test]# cat temp.lmiss CHR SNP N_MISS N_GENO F_MISS 1 snp1 3 6 0.5 1 snp2 1 6 0.1667 1 snp3 6 6 1 1 snp4 2 6 0.3333 1 snp5 0 6 0 1 snp6 0 6 0 1 snp7 0 6 0 1 snp8 3 6 0.5
