|
kmeans |
|
Scala程序 |
|
import org.apache.spark.{SparkConf, SparkContext} import org.apache.spark.mllib.clustering.{KMeans, KMeansModel} import org.apache.spark.mllib.linalg.Vectors /** * Created by hui on 2017/11/21. * K-means算法 */ object kmeans { def main(args:Array[String]): Unit ={ val conf= new SparkConf().setAppName("kmeans").setMaster("local") val sc = new SparkContext(conf) val data=sc.textFile("data/mllib/kmeans_data.txt") val parsedData=data.map(s=>Vectors.dense(s.split(' ').map(_.toDouble))).cache() val numClusters=2 val numIterations=20 val clusters=KMeans.train(parsedData,numClusters,numIterations) val WSSSE=clusters.computeCost(parsedData) println("Within Set Sum of Squared Errors = " + WSSSE) clusters.save(sc,"my_kmeans") val sameModel=KMeansModel.load(sc,"my_kmeans") } } |
|
Java程序 |
|
import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.clustering.KMeans; import org.apache.spark.mllib.clustering.KMeansModel; import org.apache.spark.mllib.linalg.Vector; import org.apache.spark.mllib.linalg.Vectors; // $example off$ public class JavaKMeansExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("JavaKMeansExample").setMaster("local"); JavaSparkContext jsc = new JavaSparkContext(conf); // $example on$ // Load and parse data String path = "data/mllib/kmeans_data.txt"; JavaRDD<String> data = jsc.textFile(path); JavaRDD<Vector> parsedData = data.map( new Function<String, Vector>() { public Vector call(String s) { String[] sarray = s.split(" "); double[] values = new double[sarray.length]; for (int i = 0; i < sarray.length; i++) { values[i] = Double.parseDouble(sarray[i]); } return Vectors.dense(values); } } ); parsedData.cache(); // Cluster the data into two classes using KMeans int numClusters = 2; int numIterations = 20; KMeansModel clusters = KMeans.train(parsedData.rdd(), numClusters, numIterations); System.out.println("Cluster centers:"); for (Vector center: clusters.clusterCenters()) { System.out.println(" " + center); } double cost = clusters.computeCost(parsedData.rdd()); System.out.println("Cost: " + cost); // Evaluate clustering by computing Within Set Sum of Squared Errors double WSSSE = clusters.computeCost(parsedData.rdd()); System.out.println("Within Set Sum of Squared Errors = " + WSSSE); // Save and load model clusters.save(jsc.sc(), "target/org/apache/spark/JavaKMeansExample/KMeansModel"); KMeansModel sameModel = KMeansModel.load(jsc.sc(), "target/org/apache/spark/JavaKMeansExample/KMeansModel"); // $example off$ jsc.stop(); } } |
|
运行结果 |
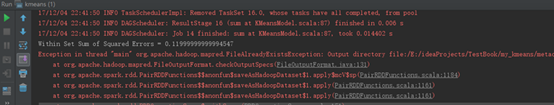 |
|
decisiontree |
|
Scala程序 |
|
import org.apache.spark.mllib.tree.DecisionTree import org.apache.spark.mllib.tree.model.DecisionTreeModel import org.apache.spark.mllib.util.MLUtils import org.apache.spark.SparkConf import org.apache.spark.SparkContext /** * Created by hui on 2017/11/21. * 使用树深为5的决策树进行分类 */ object decisiontree { def main(args:Array[String]): Unit = { val conf = new SparkConf().setAppName("decisiontree").setMaster("local") val sc = new SparkContext(conf) val data = MLUtils.loadLibSVMFile(sc, "E:\ideaProjects\TestBook\data\mllib\sample_libsvm_data.txt") val splits = data.randomSplit(Array(0.7, 0.3)) val (trainingData, testData) = (splits(0), splits(1)) val numClass = 2 val categoricalFeaturesInfo = Map[Int, Int]() val impurity = "gini" val maxDepth = 5 val maxBins = 32 val model = DecisionTree.trainClassifier(trainingData, numClass, categoricalFeaturesInfo, impurity, maxDepth, maxBins) val labelAndPreds = testData.map { point => val predicition = model.predict(point.features) (point.label, predicition) } val testErr = labelAndPreds.filter(r => r._1 != r._2).count.toDouble / testData.count() println("Test Error=" + testErr) println("Learn classification tree model: " + model.toDebugString) model.save(sc, "my_decisiontree") val sameModel = DecisionTreeModel.load(sc, "my_decisiontree") } } |
|
Java程序 |
|
import java.util.HashMap; import java.util.Map; import scala.Tuple2; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.DecisionTree; import org.apache.spark.mllib.tree.model.DecisionTreeModel; import org.apache.spark.mllib.util.MLUtils; // $example off$ class JavaDecisionTreeClassificationExample { public static void main(String[] args) { // $example on$ SparkConf sparkConf = new SparkConf().setAppName("JavaDecisionTreeClassificationExample").setMaster("local"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Integer numClasses = 2; Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<>(); String impurity = "gini"; Integer maxDepth = 5; Integer maxBins = 32; // Train a DecisionTree model for classification. final DecisionTreeModel model = DecisionTree.trainClassifier(trainingData, numClasses, categoricalFeaturesInfo, impurity, maxDepth, maxBins); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { @Override public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification tree model: " + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myDecisionTreeClassificationModel"); DecisionTreeModel sameModel = DecisionTreeModel .load(jsc.sc(), "target/tmp/myDecisionTreeClassificationModel"); // $example off$ } } |
|
运行结果 |
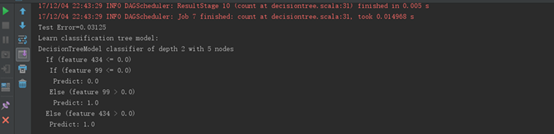 |
|
randforest_classifier |
|
Scala程序 |
|
import org.apache.spark.SparkConf import org.apache.spark.SparkContext import org.apache.spark.mllib.util.MLUtils import org.apache.spark.mllib.tree.RandomForest import org.apache.spark.mllib.tree.model.RandomForestModel /** * Created by hui on 2017/11/21. * 使用随机森林进行分类 */ object randforest_classifier { def main(args:Array[String]): Unit = { val conf = new SparkConf().setAppName("randforest_classifier").setMaster("local") val sc = new SparkContext(conf) val data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") val splits = data.randomSplit(Array(0.7, 0.3)) val (trainingData, testData) = (splits(0), splits(1)) val numClass = 2 val categoricalFeaturesInfo = Map[Int, Int]() val numTrees = 3 val featureSubsetStrategy = "auto" val impurity = "gini" val maxDepth = 4 val maxBins = 32 val model = RandomForest.trainClassifier(trainingData, numClass, categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins) val labelAndPreds = testData.map { point => val prediction = model.predict(point.features) (point.label, prediction) } val testErr = labelAndPreds.filter(r => r._1 != r._2).count.toDouble / testData.count() println("Test Error=" + testErr) println("Learned classification forest model: " + model.toDebugString) model.save(sc, "myModelPath") val sameModel = RandomForestModel.load(sc, "myModelPath") } } |
|
Java程序 |
|
import java.util.HashMap; import scala.Tuple2; import org.apache.spark.SparkConf; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.RandomForest; import org.apache.spark.mllib.tree.model.RandomForestModel; import org.apache.spark.mllib.util.MLUtils; // $example off$ public class JavaRandomForestClassificationExample { public static void main(String[] args) { // $example on$ SparkConf sparkConf = new SparkConf().setAppName("JavaRandomForestClassificationExample").setMaster("local"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Train a RandomForest model. // Empty categoricalFeaturesInfo indicates all features are continuous. Integer numClasses = 2; HashMap<Integer, Integer> categoricalFeaturesInfo = new HashMap<>(); Integer numTrees = 3; // Use more in practice. String featureSubsetStrategy = "auto"; // Let the algorithm choose. String impurity = "gini"; Integer maxDepth = 5; Integer maxBins = 32; Integer seed = 12345; final RandomForestModel model = RandomForest.trainClassifier(trainingData, numClasses, categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins, seed); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<>(model.predict(p.features()), p.label()); } }); Double testErr = 1.0 * predictionAndLabel.filter(new Function<Tuple2<Double, Double>, Boolean>() { @Override public Boolean call(Tuple2<Double, Double> pl) { return !pl._1().equals(pl._2()); } }).count() / testData.count(); System.out.println("Test Error: " + testErr); System.out.println("Learned classification forest model: " + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myRandomForestClassificationModel"); RandomForestModel sameModel = RandomForestModel.load(jsc.sc(), "target/tmp/myRandomForestClassificationModel"); // $example off$ jsc.stop(); } } |
|
运行结果 |
 |
|
randforest_regressor |
|
Scala程序 |
|
import org.apache.spark.{SparkConf, SparkContext} import org.apache.spark.mllib.util.MLUtils import org.apache.spark.SparkConf import org.apache.spark.SparkContext import org.apache.spark.mllib.util.MLUtils import org.apache.spark.mllib.tree.RandomForest import org.apache.spark.mllib.tree.model.RandomForestModel /** * Created by hui on 2017/11/21. * 使用随机森林进行回归 */ object randforest_regressor { def main(args:Array[String]): Unit = { val conf = new SparkConf().setAppName("randforest_regressor").setMaster("local") val sc = new SparkContext(conf) val data = MLUtils.loadLibSVMFile(sc, "data/mllib/sample_libsvm_data.txt") val splits = data.randomSplit(Array(0.7, 0.3)) val (trainingData, testData) = (splits(0), splits(1)) val numClass = 2 val categoricalFeaturesInfo = Map[Int, Int]() val numTrees = 3 val featureSubsetStrategy = "auto" val impurity = "variance" val maxDepth = 4 val maxBins = 32 val model = RandomForest.trainRegressor(trainingData, categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins) val labelAndPredictions = testData.map { point => val prediction = model.predict(point.features) (point.label, prediction) } val testMSE = labelAndPredictions.map { case (v, p) => math.pow((v - p), 2) }.mean() println("Test Mean Squared Error=" + testMSE) println("Learned regression forest model: " + model.toDebugString) model.save(sc, "myModelPath") val sameModel = RandomForestModel.load(sc, "myModelPath") } } |
|
Java程序 |
|
import java.util.HashMap; import java.util.Map; import scala.Tuple2; import org.apache.spark.api.java.function.Function2; import org.apache.spark.api.java.JavaPairRDD; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.JavaSparkContext; import org.apache.spark.api.java.function.Function; import org.apache.spark.api.java.function.PairFunction; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.tree.RandomForest; import org.apache.spark.mllib.tree.model.RandomForestModel; import org.apache.spark.mllib.util.MLUtils; import org.apache.spark.SparkConf; // $example off$ public class JavaRandomForestRegressionExample { public static void main(String[] args) { // $example on$ SparkConf sparkConf = new SparkConf().setAppName("JavaRandomForestRegressionExample").setMaster("local"); JavaSparkContext jsc = new JavaSparkContext(sparkConf); // Load and parse the data file. String datapath = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(jsc.sc(), datapath).toJavaRDD(); // Split the data into training and test sets (30% held out for testing) JavaRDD<LabeledPoint>[] splits = data.randomSplit(new double[]{0.7, 0.3}); JavaRDD<LabeledPoint> trainingData = splits[0]; JavaRDD<LabeledPoint> testData = splits[1]; // Set parameters. // Empty categoricalFeaturesInfo indicates all features are continuous. Map<Integer, Integer> categoricalFeaturesInfo = new HashMap<>(); Integer numTrees = 3; // Use more in practice. String featureSubsetStrategy = "auto"; // Let the algorithm choose. String impurity = "variance"; Integer maxDepth = 4; Integer maxBins = 32; Integer seed = 12345; // Train a RandomForest model. final RandomForestModel model = RandomForest.trainRegressor(trainingData, categoricalFeaturesInfo, numTrees, featureSubsetStrategy, impurity, maxDepth, maxBins, seed); // Evaluate model on test instances and compute test error JavaPairRDD<Double, Double> predictionAndLabel = testData.mapToPair(new PairFunction<LabeledPoint, Double, Double>() { @Override public Tuple2<Double, Double> call(LabeledPoint p) { return new Tuple2<>(model.predict(p.features()), p.label()); } }); Double testMSE = predictionAndLabel.map(new Function<Tuple2<Double, Double>, Double>() { @Override public Double call(Tuple2<Double, Double> pl) { Double diff = pl._1() - pl._2(); return diff * diff; } }).reduce(new Function2<Double, Double, Double>() { @Override public Double call(Double a, Double b) { return a + b; } }) / testData.count(); System.out.println("Test Mean Squared Error: " + testMSE); System.out.println("Learned regression forest model: " + model.toDebugString()); // Save and load model model.save(jsc.sc(), "target/tmp/myRandomForestRegressionModel"); RandomForestModel sameModel = RandomForestModel.load(jsc.sc(), "target/tmp/myRandomForestRegressionModel"); // $example off$ jsc.stop(); } } |
|
运行结果 |
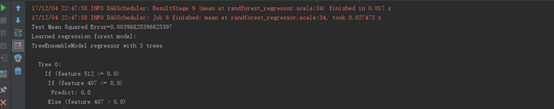 |
|
svm |
|
Scala程序 |
|
import org.apache.spark.{SparkConf, SparkContext} import org.apache.spark.mllib.classification.{SVMModel, SVMWithSGD} import org.apache.spark.mllib.evaluation.BinaryClassificationMetrics import org.apache.spark.mllib.util.MLUtils /** * Created by hui on 2017/11/21. * 支持向量机分类 */ object svm { def main(args:Array[String]): Unit = { val conf = new SparkConf().setAppName("svm").setMaster("local") val sc = new SparkContext(conf) val data=MLUtils.loadLibSVMFile(sc,"data/mllib/sample_libsvm_data.txt") val splits=data.randomSplit(Array(0.6,0.4),seed=11L) val training=splits(0).cache() val test=splits(1) val numIterations=100 val model=SVMWithSGD.train(training,numIterations) model.clearThreshold() val scoreAndLabels=test.map{point=> val score=model.predict(point.features) (score,point.label) } val metrics=new BinaryClassificationMetrics(scoreAndLabels) val auROC=metrics.areaUnderROC() println("Area under ROC="+ auROC) model.save(sc,"my_svm") val sameModel=SVMModel.load(sc,"my_svm") } } |
|
Java程序 |
|
import org.apache.spark.SparkConf; import org.apache.spark.SparkContext; // $example on$ import scala.Tuple2; import org.apache.spark.api.java.JavaRDD; import org.apache.spark.api.java.function.Function; import org.apache.spark.mllib.classification.SVMModel; import org.apache.spark.mllib.classification.SVMWithSGD; import org.apache.spark.mllib.evaluation.BinaryClassificationMetrics; import org.apache.spark.mllib.regression.LabeledPoint; import org.apache.spark.mllib.util.MLUtils; // $example off$ /** * Example for SVMWithSGD. */ public class JavaSVMWithSGDExample { public static void main(String[] args) { SparkConf conf = new SparkConf().setAppName("JavaSVMWithSGDExample").setMaster("local"); SparkContext sc = new SparkContext(conf); // $example on$ String path = "data/mllib/sample_libsvm_data.txt"; JavaRDD<LabeledPoint> data = MLUtils.loadLibSVMFile(sc, path).toJavaRDD(); // Split initial RDD into two... [60% training data, 40% testing data]. JavaRDD<LabeledPoint> training = data.sample(false, 0.6, 11L); training.cache(); JavaRDD<LabeledPoint> test = data.subtract(training); // Run training algorithm to build the model. int numIterations = 100; final SVMModel model = SVMWithSGD.train(training.rdd(), numIterations); // Clear the default threshold. model.clearThreshold(); // Compute raw scores on the test set. JavaRDD<Tuple2<Object, Object>> scoreAndLabels = test.map( new Function<LabeledPoint, Tuple2<Object, Object>>() { public Tuple2<Object, Object> call(LabeledPoint p) { Double score = model.predict(p.features()); return new Tuple2<Object, Object>(score, p.label()); } } ); // Get evaluation metrics. BinaryClassificationMetrics metrics = new BinaryClassificationMetrics(JavaRDD.toRDD(scoreAndLabels)); double auROC = metrics.areaUnderROC(); System.out.println("Area under ROC = " + auROC); // Save and load model model.save(sc, "target/tmp/javaSVMWithSGDModel"); SVMModel sameModel = SVMModel.load(sc, "target/tmp/javaSVMWithSGDModel"); // $example off$ sc.stop(); } } |
|
运行结果 |
 |