[root@ibiomed ~]# cat > test.sh
#!/bin/bash
## function: split file by specified line count
## parameter 1: file name to be splitted
## parameter 2: line number of new files
filename=$1
fileline=$2
echo "filename=$filename"
echo "fileline=$fileline"
awk -v count=$fileline 'BEGIN{i=0} { print $0 > sprintf("%s_%d",FILENAME,i) ; if (NR>=(i+1)*count) { close(sprintf("%s_%d",FILENAME,i)); i++;} }' $filename
echo "=====finish====="
[root@ibiomed ~]# cat > temp.txt >ACTL6A_1 GGATAGTTTCCAAGCTATTT >ACTL6A_3 TTTGCTAATGGTCGTTCTAC >ACTL6A_5 GTTGAAGGACATAGCCATCG >ACTL6A_7 ACTGCAATTCCAGTCCACGA
[root@ibiomed ~]# chmod 777 test.sh
[root@ibiomed ~]# ./test.sh temp.txt 2
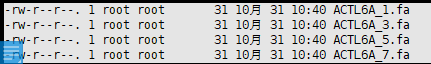
[root@ibiomed ~]# more temp.txt_0 >ACTL6A_1 GGATAGTTTCCAAGCTATTT
This works. New files with new file names, i.e. :
#!/bin/bash
## function: split file by specified line count
## parameter 1: file name to be splitted
## parameter 2: line number of new files
filename=$1
fileline=$2
echo "filename=$filename"
echo "fileline=$fileline"
#awk -v count=$fileline 'BEGIN{i=0} { print $0 > sprintf("%s_%d",FILENAME,i) ; if (NR>=(i+1)*count) { close(sprintf("%s_%d",FILENAME,i)); i++;} }' $filename
awk -v count=$fileline 'BEGIN{i=0; NEWFILE="test"} { if (NR<(i+1)*count) { NEWFILE=gensub(/>/,"",1,$0);} print $0 > sprintf("%s.fa",NEWFILE) ; if (NR>=(i+1)*count) { close(sprintf("%s.fa",NEWFILE)); i++;} }' $filename
echo "=====finish====="
REF
https://www.cnblogs.com/cyttina/p/7471221.html