这篇文章
https://zhuanlan.zhihu.com/p/250146007
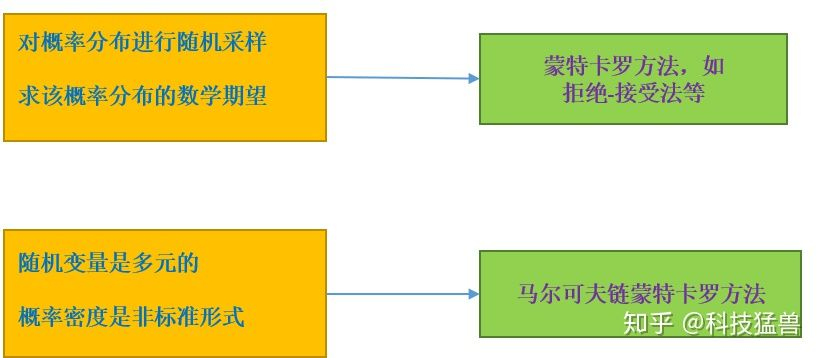
其实随机算法分为两类:蒙特卡罗方法和拉斯维加斯方法,蒙特卡罗方法指的是算法的时间复杂度固定,然而结果有一定几率失败,采样越多结果越好。拉斯维加斯方法指的是算法一定成功,然而运行时间是概率的。
- 不可约:每个状态都能去到。
- 非周期:返回时间公约数是1。
- 正常返:离开此状态有限步一定能回来。迟早会回来。
- 零常返:离开此状态能回来,但需要无穷多步。
- 非常返:离开此状态有限步不一定回得来。
- 遍历定理:不可约,非周期,正常返
有唯一的平稳分布。

常用的马尔可夫链蒙特卡罗法有Metropolis-Hastings算法、吉布斯抽样。
MCMC算法的一般流程是:先给定目标分布完成采样过程,若目标分布是一维的,就用M-H采样方法;若目标分布是多维的,就用Gibbs采样方法。采样结束之后,蒙特卡罗方法来用样本集模拟求和,求出目标变量(期望等)的统计值作为估计值。这套思路被应用于概率分布的估计、定积分的近似计算、最优化问题的近似求解等问题,特别是被应用于统计学习中概率模型的学习与推理,是重要的统计学习计算方法。
下面这篇文章讲得也挺好,我觉得代码部分比上面的好一些
https://blog.csdn.net/weixin_39953236/article/details/111369770?spm=1001.2101.3001.6650.7&utm_medium=distribute.pc_relevant.none-task-blog-2%7Edefault%7EBlogCommendFromBaidu%7Edefault-7.pc_relevant_default&depth_1-utm_source=distribute.pc_relevant.none-task-blog-2%7Edefault%7EBlogCommendFromBaidu%7Edefault-7.pc_relevant_default&utm_relevant_index=10
MH算法
# -*- coding:utf-8 -*- import random import numpy as np import matplotlib.pyplot as plt def mh(q, p, m, n): # randomize a number x = random.uniform(0.1, 1) for t in range(0, m+n): x_sample = q.sample(x) try: accept_prob = min(1, p.prob(x_sample)*q.prob(x_sample, x)/(p.prob(x)*q.prob(x, x_sample))) except: accept_prob = 0 u = random.uniform(0, 1) if u < accept_prob: x = x_sample if t >= m: yield x class Exponential(object): def __init__(self, scale): self.scale = scale self.lam = 1.0 / scale def prob(self, x): if x <= 0: raise Exception("The sample shouldn't be less than zero") result = self.lam * np.exp(-x * self.lam) return result def sample(self, num): sample = np.random.exponential(self.scale, num) return sample # 假设我们的目标概率密度函数p1(x)是指数概率密度函数 scale = 5 p1 = Exponential(scale) class Norm(): def __init__(self, mean, std): self.mean = mean self.sigma = std def prob(self, x): return np.exp(-(x - self.mean) ** 2 / (2 * self.sigma ** 2.0)) * 1.0 / (np.sqrt(2 * np.pi) * self.sigma) def sample(self, num): sample = np.random.normal(self.mean, self.sigma, size=num) return sample # 假设我们的目标概率密度函数p1(x)是均值方差分别为3,2的正态分布 p2 = Norm(3, 2) class Transition(): def __init__(self, sigma): self.sigma = sigma def sample(self, cur_mean): cur_sample = np.random.normal(cur_mean, scale=self.sigma, size=1)[0] return cur_sample def prob(self, mean, x): return np.exp(-(x-mean)**2/(2*self.sigma**2.0)) * 1.0/(np.sqrt(2 * np.pi)*self.sigma) # 假设我们的转移核方差为10的正态分布 q = Transition(10) m = 100 n = 100000 # 采样个数 simulate_samples_p1 = [li for li in mh(q, p1, m, n)] plt.subplot(2,2,1) plt.hist(simulate_samples_p1, 100) plt.title("Simulated X ~ Exponential(1/5)") samples = p1.sample(n) plt.subplot(2,2,2) plt.hist(samples, 100) plt.title("True X ~ Exponential(1/5)") simulate_samples_p2 = [li for li in mh(q, p2, m, n)] plt.subplot(2,2,3) plt.hist(simulate_samples_p2, 50) plt.title("Simulated X ~ N(3,2)") samples = p2.sample(n) plt.subplot(2,2,4) plt.hist(samples, 50) plt.title("True X ~ N(3,2)") plt.suptitle("Transition Kernel N(0,10)simulation results") plt.show()
Gibbs采样
import matplotlib.pyplot as plt from mpl_toolkits.mplot3d import Axes3D import numpy as np class Transition(): def __init__(self, mean, cov): self.mean = mean self.sigmas = [] for i in range(K): self.sigmas.append(np.sqrt(cov[i][i])) self.rho = cov[0][1]/(self.sigmas[0] * self.sigmas[1]) def sample(self, id1, id2_list, x2_list): id2 = id2_list[0] # only consider two dimension x2 = x2_list[0] # only consider two dimension cur_mean = self.mean[id1] + self.rho*self.sigmas[id1]/self.sigmas[id2] * (x2-self.mean[id2]) cur_sigma = (1-self.rho**2) * self.sigmas[id1]**2 return np.random.normal(cur_mean, scale=cur_sigma, size=1)[0] def gibbs(p, m, n): # randomize a number x = np.random.rand(K) for t in range(0, m+n): for j in range(K): total_indexes = list(range(K)) total_indexes.remove(j) left_x = x[total_indexes] x[j] = p.sample(j, total_indexes, left_x) if t >= m: yield x mean = [5, 8] cov = [[1, 0.5], [0.5, 1]] K = len(mean) q = Transition(mean, cov) m = 100 n = 1000 gib = gibbs(q, m, n) simulated_samples = [] x_samples = [] y_samples = [] for li in gib: x_samples.append(li[0]) y_samples.append(li[1]) fig = plt.figure() ax = fig.add_subplot(131, projection='3d') hist, xedges, yedges = np.histogram2d(x_samples, y_samples, bins=100, range=[[0,10],[0,16]]) xpos, ypos = np.meshgrid(xedges[:-1], yedges[:-1]) xpos = xpos.ravel() ypos = ypos.ravel() zpos = 0 dx = xedges[1] - xedges[0] dy = yedges[1] - yedges[0] dz = hist.flatten() ax.bar3d(xpos, ypos, zpos, dx, dy, dz, zsort='average') ax = fig.add_subplot(132) ax.hist(x_samples, bins=50) ax.set_title("Simulated on dim1") ax = fig.add_subplot(133) ax.hist(y_samples, bins=50) ax.set_title("Simulated on dim2") plt.show()