iris数据集预测(对比随机森林和逻辑回归算法)
随机森林
library(randomForest)
#挑选响应变量
index <- subset(iris,Species != "setosa")
ir <- droplevels(index)
set.seed(1)
ind<-sample(2,nrow(ir),replace=TRUE,prob=c(0.7,0.3))
train<-ir[ind==1,]
test<-ir[ind==2,]
rf<-randomForest(Species~.,data=train,ntree=100)
rf
Call:
randomForest(formula = Species ~ ., data = train, ntree = 100)
Type of random forest: classification
Number of trees: 100
No. of variables tried at each split: 2
OOB estimate of error rate: 5.88%
Confusion matrix:
versicolor virginica class.error
versicolor 32 2 0.05882353
virginica 2 32 0.05882353
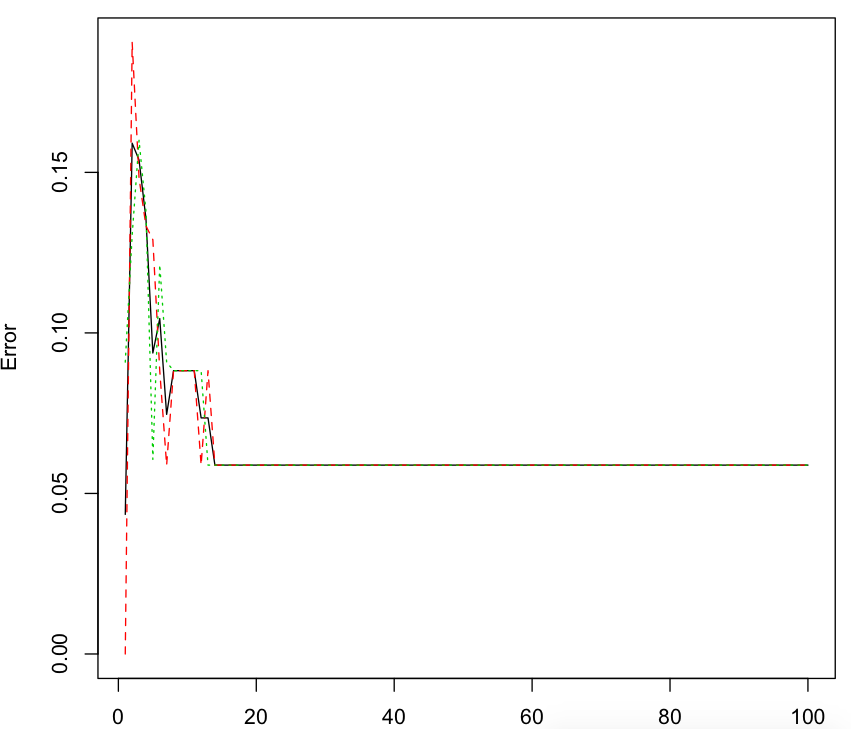
#随机森林的误差率
plot(rf)
#变量重要性
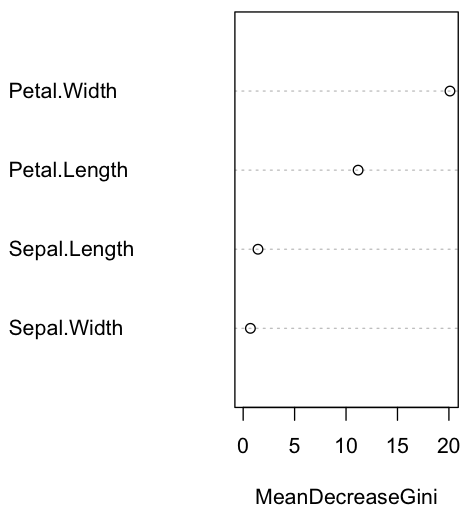
importance(rf)
importance(rf)
MeanDecreaseGini
Sepal.Length 1.4398647
Sepal.Width 0.7037353
Petal.Length 11.1734509
Petal.Width 20.1025569
varImpPlot(rf)
#查看预测结果
pred<-predict(rf,newdata=test)
table(pred,test$Species) pred versicolor virginica
versicolor 15 2
virginica 1 14
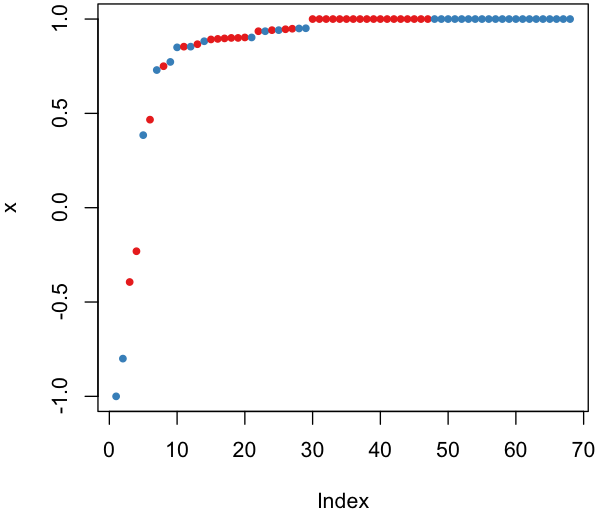
#预测边距
plot(margin(rf,test$Species))
逻辑回归
library(pROC)
g1<-glm(Species~.,family=binomial(link='logit'),data=train)
pre1<-predict(g1,type="response")
g1 Call: glm(formula = Species ~ ., family = binomial(link = "logit"), data = train) Coefficients: (Intercept) Sepal.Length Sepal.Width Petal.Length Petal.Width -32.01349 -3.85855 -0.02084 6.65355 14.08817 Degrees of Freedom: 67 Total (i.e. Null); 63 Residual Null Deviance: 94.27 Residual Deviance: 8.309 AIC: 18.31
summary(g1)
Call:
glm(formula = Species ~ ., family = binomial(link = "logit"),
data = train)
Deviance Residuals:
Min 1Q Median 3Q Max
-1.73457 -0.02241 -0.00011 0.03691 1.76243
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -32.01349 28.51193 -1.123 0.2615
Sepal.Length -3.85855 3.16430 -1.219 0.2227
Sepal.Width -0.02084 4.85883 -0.004 0.9966
Petal.Length 6.65355 5.47953 1.214 0.2246
Petal.Width 14.08817 7.32507 1.923 0.0544 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 94.268 on 67 degrees of freedom
Residual deviance: 8.309 on 63 degrees of freedom
AIC: 18.309
Number of Fisher Scoring iterations: 9
#方差分析
anova(g1,test="Chisq") Analysis of Deviance Table Model: binomial, link: logit Response: Species Terms added sequentially (first to last) Df Deviance Resid. Df Resid. Dev Pr(>Chi) NULL 67 94.268 Sepal.Length 1 14.045 66 80.223 0.0001785 *** Sepal.Width 1 0.782 65 79.441 0.3764212 Petal.Length 1 62.426 64 17.015 2.766e-15 *** Petal.Width 1 8.706 63 8.309 0.0031715 ** --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#计算最优阀值
modelroc1<-roc(as.factor(ifelse(train$Species=="virginica",1,0)),pre1)
plot(modelroc1,print.thres=TRUE)

评估模型的预测效果
predict <-predict(g1,type="response",newdata=test)
predict.results <-ifelse(predict>0.804,"virginica","versicolor")
misClasificError <-mean(predict.results !=test$Species)
print(paste("Accuracy:",1-misClasificError)) [1] "Accuracy: 0.90625"
XGBoost
y<-data.matrix(as.data.frame(train$Species))-1
x<-data.matrix(train[-5])
bst <- xgboost(data =x, label = y, max.depth = 2, eta = 1,nround = 2, objective = "binary:logistic") [1] train-error:0.029412 [2] train-error:0.029412
p<-predict(bst,newdata=data.matrix(test))
modelroc2<-roc(as.factor(ifelse(test$Species=="virginica",1,0)),p)
plot(modelroc2)

predict.results <-ifelse(p>0.11,"virginica","versicolor")
misClasificError <-mean(predict.results !=test$Species)
print(paste(1-misClasificError)) [1] "0.90625"