原创转载请注明出处:https://www.cnblogs.com/agilestyle/p/12855528.html
特征缩放
归一化
将一列数据变化到某个固定区间(范围)中,通常这个区间是[0, 1]
标准化
将数据变换为均值为0,标准差为1的分布
Rescaling (min-max normalization)
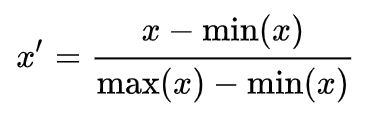
Mean normalization
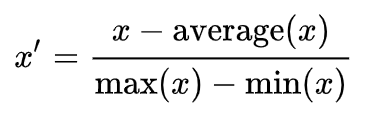
Standardization (Z-score Normalization)
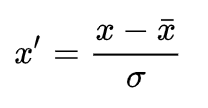
Scaling to unit length
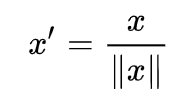
使用 MinMaxScaler 归一化
参考 min-max normalization 数学公式 进行 MinMaxScaler
import numpy as np from sklearn.preprocessing import MinMaxScaler X = np.array([[1, 2000], [2, 3000], [3, 4000], [4, 5000], [5, 1000]], dtype='float') # (5, 2) print(X.shape) # [[1.e+00 2.e+03] # [2.e+00 3.e+03] # [3.e+00 4.e+03] # [4.e+00 5.e+03] # [5.e+00 1.e+03]] print(X) # max(x) X_max = np.max(X, axis=0) # X_max: [ 5. 5000.] print("X_max: ", X_max) # X_min: [ 1. 1000.] X_min = np.min(X, axis=0) print("X_min: ", X_min) # max(x) - min(x) scope = X_max - X_min # scope: [ 4. 4000.] print("scope: ", scope) # mean normalization mean = np.mean(X, axis=0) # [ 3. 3000.] print(mean) mn_X = (X - mean) / (X_max - X_min) # mean normalization: # [[-0.5 -0.25] # [-0.25 0. ] # [ 0. 0.25] # [ 0.25 0.5 ] # [ 0.5 -0.5 ]] print("mean normalization: ", mn_X) # min-max normalization manual_mms_X = (X - X_min) / (X_max - X_min) # mms_X: # [[0. 0.25] # [0.25 0.5 ] # [0.5 0.75] # [0.75 1. ] # [1. 0. ]] print("mms_X: ", manual_mms_X) mms = MinMaxScaler() mms_X = mms.fit_transform(X) # mms_X: # [[0. 0.25] # [0.25 0.5 ] # [0.5 0.75] # [0.75 1. ] # [1. 0. ]] print("mms_X: ", mms_X)
Note:
- MinMaxScaler:归一到 [0,1]
- MaxAbsScaler:归一到 [-1,1]
使用 PCA 将 mms_X 从二维降到一维
import numpy as np from sklearn.preprocessing import MinMaxScaler X = np.array([[1, 2000], [2, 3000], [3, 4000], [4, 5000], [5, 1000]], dtype='float') # (5, 2) print(X.shape) # [[1.e+00 2.e+03] # [2.e+00 3.e+03] # [3.e+00 4.e+03] # [4.e+00 5.e+03] # [5.e+00 1.e+03]] print(X) # max(x) X_max = np.max(X, axis=0) # X_max: [ 5. 5000.] print("X_max: ", X_max) # X_min: [ 1. 1000.] X_min = np.min(X, axis=0) print("X_min: ", X_min) # max(x) - min(x) scope = X_max - X_min # scope: [ 4. 4000.] print("scope: ", scope) # mean normalization mean = np.mean(X, axis=0) # [ 3. 3000.] print(mean) mn_X = (X - mean) / (X_max - X_min) # mean normalization: # [[-0.5 -0.25] # [-0.25 0. ] # [ 0. 0.25] # [ 0.25 0.5 ] # [ 0.5 -0.5 ]] print("mean normalization: ", mn_X) # min-max normalization manual_mms_X = (X - X_min) / (X_max - X_min) # mms_X: # [[0. 0.25] # [0.25 0.5 ] # [0.5 0.75] # [0.75 1. ] # [1. 0. ]] print("mms_X: ", manual_mms_X) mms = MinMaxScaler() mms_X = mms.fit_transform(X) # mms_X: # [[0. 0.25] # [0.25 0.5 ] # [0.5 0.75] # [0.75 1. ] # [1. 0. ]] print("mms_X: ", mms_X) from sklearn.decomposition import PCA pca = PCA(n_components=1) # PCA(copy=True, iterated_power='auto', n_components=1, random_state=None, # svd_solver='auto', tol=0.0, whiten=False) print(pca) # pca.fit(mms_X) pca.fit(manual_mms_X) # pca_X = pca.transform(mms_X) pca_X = pca.transform(manual_mms_X) # 2 dimensions to 1 dimension: # [[ 0.5 ] # [ 0.25] # [ 0. ] # [-0.25] # [-0.5 ]] print("2 dimensions to 1 dimension: ", pca_X) # 代表降维后的各主成分的方差值,方差值越大,则说明越是重要的主成分 # explained_variance_: [0.15625] print("explained_variance_:", pca.explained_variance_) # 代表降维后的各主成分的方差值占总方差值的比例,这个比例越大,则越是重要的主成分 # explained_variance_ratio_: [0.5] print("explained_variance_ratio_:", pca.explained_variance_ratio_) # 最终确定的主成分 # components_: [[-1. -0.]] print("components_: ", pca.components_) inverse_pca_X = pca.inverse_transform(pca_X) # inverse_transform: # [[0. 0.5 ] # [0.25 0.5 ] # [0.5 0.5 ] # [0.75 0.5 ] # [1. 0.5 ]] print("inverse_transform: ", inverse_pca_X)
使用 PCA 从三维降到二维
准备数据
import matplotlib.pyplot as plt from mpl_toolkits.mplot3d import Axes3D from sklearn.datasets import make_blobs # X为样本特征,Y为样本簇类别, 共10000个样本,每个样本3个特征,共4个簇 X, y = make_blobs(n_samples=10000, n_features=3, centers=[[3, 3, 3], [0, 0, 0], [1, 1, 1], [2, 2, 2]], cluster_std=[0.2, 0.1, 0.2, 0.2], random_state=9) # (10000, 3) print(X.shape) # (10000,) print(y.shape) fig = plt.figure() ax = Axes3D(fig) ax.scatter(X[:, 0], X[:, 1], X[:, 2], marker='o', c=y) ax.set_xlabel('X') ax.set_ylabel('Y') ax.set_zlabel('Z') plt.show()
三维数据分布图如下:
先不降维,只对数据进行投影,看看投影后的三个维度的方差分布
from sklearn.decomposition import PCA # 先不降维,只对数据进行投影 pca = PCA(n_components=3) pca.fit(X) # [0.98318212 0.00850037 0.00831751] print(pca.explained_variance_ratio_) # [3.78521638 0.03272613 0.03202212] print(pca.explained_variance_)
Note: 可以看出投影后三个特征维度的方差比例大约为98.3%:0.8%:0.8%。投影后第一个特征占了绝大多数的主成分比例。
进行降维,从三维降到二维
# 降维,从三维降到二维 pca = PCA(n_components=2) pca.fit(X) # [0.98318212 0.00850037] print(pca.explained_variance_ratio_) # [3.78521638 0.03272613] print(pca.explained_variance_)
Note: 结果其实可以预料,因为上面三个投影后的特征维度的方差分别为:[ 3.78483785 0.03272285 0.03201892],投影到二维后选择的肯定是前两个特征,而抛弃第三个特征。
查看此时转化后的数据分布
X_new = pca.transform(X) plt.scatter(X_new[:, 0], X_new[:, 1], marker='o', c=y) plt.show()
二维数据分布图如下:

不直接指定降维的维度,而指定降维后的主成分方差和比例,比如指定了主成分至少占95%
# 指定了主成分至少占95% pca = PCA(n_components=0.95) pca.fit(X) # [0.98318212] print(pca.explained_variance_ratio_) # [3.78521638] print(pca.explained_variance_) # 1 print(pca.n_components_)
Note: 只有第一个投影特征被保留。这也很好理解,第一个主成分占投影特征的方差比例高达98%。只选择这一个特征维度便可以满足95%的阈值。
指定了主成分至少占99%
# 指定了主成分至少占99% pca = PCA(n_components=0.99) pca.fit(X) # [0.98318212 0.00850037] print(pca.explained_variance_ratio_) # [3.78521638 0.03272613] print(pca.explained_variance_) # 2 print(pca.n_components_)
Note: 第一个主成分占了98.3%的方差比例,第二个主成分占了0.8%的方差比例,两者一起可以满足99%的阈值。
让MLE算法自己选择降维维度的效果
# 让MLE算法自己选择降维维度 pca = PCA(n_components='mle') pca.fit(X) # [0.98318212] print(pca.explained_variance_ratio_) # [3.78521638] print(pca.explained_variance_) # 1 print(pca.n_components_)
Note: 数据的第一个投影特征的方差占比高达98.3%,MLE算法只保留了第一个特征
Reference
https://en.wikipedia.org/wiki/Feature_scaling
https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html
https://scikit-learn.org/stable/modules/preprocessing.html
https://scikit-learn.org/stable/modules/generated/sklearn.decomposition.PCA.html