Non-bonded interactions
NAMD has a number of options that control the way that non-bonded interactions are calculated. These options are interrelated and can be quite confusing, so this section attempts to explain the behavior of the non-bonded interactions and how to use these parameters.
Van der Waals interactions
The simplest non-bonded interaction is the van der Waals interaction. In NAMD, van der Waals interactions are always truncated at the cutoff distance, specified by cutoff. The main option that effects van der Waals interactions is the switching parameter. With this option set to on, a smooth switching function will be used to truncate the van der Waals potential energy smoothly at the cutoff distance. A graph of the van der Waals potential with this switching function is shown in Figure 1. If switching is set to off, the van der Waals energy is just abruptly truncated at the cutoff distance, so that energy may not be conserved.
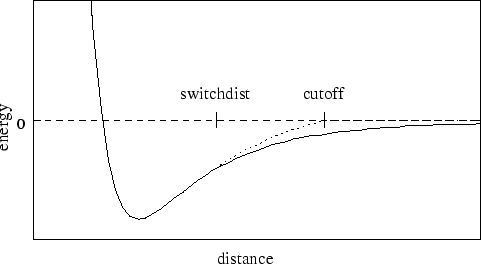 |
The switching function used is based on the X-PLOR switching function. The parameter switchdist specifies the distance at which the switching function should start taking effect to bring the van der Waals potential to 0 smoothly at the cutoff distance. Thus, the value of switchdist must always be less than that of cutoff.
Electrostatic interactions
The handling of electrostatics is slightly more complicated due to the incorporation of multiple timestepping for full electrostatic interactions. There are two cases to consider, one where full electrostatics is employed and the other where electrostatics are truncated at a given distance.
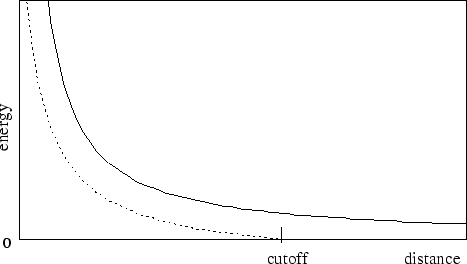
First let us consider the latter case, where electrostatics are truncated at the cutoff distance. Using this scheme, all electrostatic interactions beyond a specified distance are ignored, or assumed to be zero. If switching is set toon, rather than having a discontinuity in the potential at the cutoff distance, a shifting function is applied to the electrostatic potential as shown in Figure 2. As this figure shows, the shifting function shifts the entire potential curve so that the curve intersects the x-axis at the cutoff distance. This shifting function is based on the shifting function used by X-PLOR.
 |
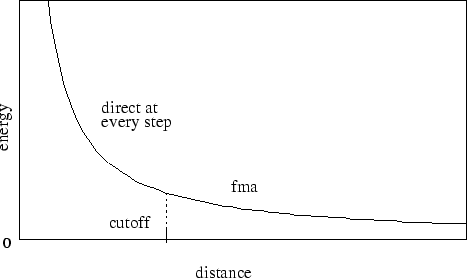
Next, consider the case where full electrostatics are calculated. In this case, the electrostatic interactions are not truncated at any distance. In this scheme, the cutoff parameter has a slightly different meaning for the electrostatic interactions -- it represents the local interaction distance, or distance within which electrostatic pairs will be directly calculated every timestep. Outside of this distance, interactions will be calculated only periodically. These forces will be applied using a multiple timestep integration scheme as described in Section 7.3.4.
 |
Non-bonded force field parameters
- cutoff
 local interaction distance common to both electrostatic and van der Waals calculations (Å)
local interaction distance common to both electrostatic and van der Waals calculations (Å) 
Acceptable Values: positive decimal
Description: See Section 5.2 for more information. - switching
 use switching function?
use switching function? 
Acceptable Values: on or off
Default Value: on
Description: If switching is specified to be off, then a truncated cutoff is performed. If switching is turned on, then smoothing functions are applied to both the electrostatics and van der Waals forces. For a complete description of the non-bonded force parameters see Section 5.2. If switching is set to on, then switchdist must also be defined. - vdwForceSwitching
 use force switching for VDW?
use force switching for VDW? 
Acceptable Values: on or off
Default Value: off
Description: If both switching and vdwForceSwitching are set to on, then CHARMM force switching is used for van der Waals forces. LJcorrection as implemented is inconsistent with vdwForceSwitching. - switchdist
 distance at which to activate switching/splitting function for electrostatic and van der Waals calculations (Å)
distance at which to activate switching/splitting function for electrostatic and van der Waals calculations (Å) 
Acceptable Values: positive decimal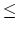 cutoff
cutoff
Description: Distance at which the switching function should begin to take effect. This parameter only has meaning if switching is set to on. The value of switchdist must be less than or equal to the value of cutoff, since the switching function is only applied on the range from switchdist to cutoff. For a complete description of the non-bonded force parameters see Section 5.2. - exclude
 non-bonded exclusion policy to use
non-bonded exclusion policy to use 
Acceptable Values: none, 1-2, 1-3, 1-4, or scaled1-4
Description: This parameter specifies which pairs of bonded atoms should be excluded from non-bonded interactions. With the value of none, no bonded pairs of atoms will be excluded. With the value of 1-2, all atom pairs that are directly connected via a linear bond will be excluded. With the value of 1-3, all 1-2 pairs will be excluded along with all pairs of atoms that are bonded to a common third atom (i.e., if atom A is bonded to atom B and atom B is bonded to atom C, then the atom pair A-C would be excluded). With the value of 1-4, all 1-3 pairs will be excluded along with all pairs connected by a set of two bonds (i.e., if atom A is bonded to atom B, and atom B is bonded to atom C, and atom C is bonded to atom D, then the atom pair A-D would be excluded). With the value of scaled1-4, all 1-3 pairs are excluded and all pairs that match the 1-4 criteria are modified. The electrostatic interactions for such pairs are modified by the constant factor defined by 1-4scaling. The van der Waals interactions are modified by using the special 1-4 parameters defined in the parameter files. The value of scaled1-4 is necessary to enable the modified 1-4 VDW parameters present in the CHARMM parameter files. - 1-4scaling
 scaling factor for 1-4 electrostatic interactions
scaling factor for 1-4 electrostatic interactions 
Acceptable Values: 0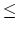 decimal
decimal 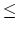 1
1
Default Value: 1.0
Description: Scaling factor for 1-4 electrostatic interactions. This factor is only used when the exclude parameter is set to scaled1-4. In this case, this factor is used to modify the electrostatic interactions between 1-4 atom pairs. If the exclude parameter is set to anything but scaled1-4, this parameter has no effect regardless of its value. - dielectric
 dielectric constant for system
dielectric constant for system 
Acceptable Values: decimal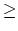 1.0
1.0
Default Value: 1.0
Description: Dielectric constant for the system. A value of 1.0 implies no modification of the electrostatic interactions. Any larger value will lessen the electrostatic forces acting in the system. - nonbondedScaling
 scaling factor for nonbonded forces
scaling factor for nonbonded forces 
Acceptable Values: decimal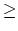 0.0
0.0
Default Value: 1.0
Description: Scaling factor for electrostatic and van der Waals forces. A value of 1.0 implies no modification of the interactions. Any smaller value will lessen the nonbonded forces acting in the system. - vdwGeometricSigma
 use geometric mean to combine L-J sigmas
use geometric mean to combine L-J sigmas 
Acceptable Values: yes or no
Default Value: no
Description: Use geometric mean, as required by OPLS, rather than traditional arithmetic mean when combining Lennard-Jones sigma parameters for different atom types. - limitdist
 maximum distance between pairs for limiting interaction strength(Å)
maximum distance between pairs for limiting interaction strength(Å) 
Acceptable Values: non-negative decimal
Default Value: 0.
Description: The electrostatic and van der Waals potential functions diverge as the distance between two atoms approaches zero. The potential for atoms closer than limitdist is instead treated as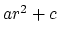 with parameters chosen to match the force and potential at limitdist. This option should primarily be useful for alchemical free energy perturbation calculations, since it makes the process of creating and destroying atoms far less drastic energetically. The larger the value of limitdist the more the maximum force between atoms will be reduced. In order to not alter the other interactions in the simulation, limitdist should be less than the closest approach of any non-bonded pair of atoms; 1.3Å appears to satisfy this for typical simulations but the user is encouraged to experiment. There should be no performance impact from enabling this feature.
with parameters chosen to match the force and potential at limitdist. This option should primarily be useful for alchemical free energy perturbation calculations, since it makes the process of creating and destroying atoms far less drastic energetically. The larger the value of limitdist the more the maximum force between atoms will be reduced. In order to not alter the other interactions in the simulation, limitdist should be less than the closest approach of any non-bonded pair of atoms; 1.3Å appears to satisfy this for typical simulations but the user is encouraged to experiment. There should be no performance impact from enabling this feature.
- LJcorrection
 Apply long-range corrections to the system energy and virial to account for neglected vdW forces?
Apply long-range corrections to the system energy and virial to account for neglected vdW forces? 
Acceptable Values: yes or no
Default Value: no
Description: Apply an analytical correction to the reported vdW energy and virial that is equal to the amount lost due to switching and cutoff of the LJ potential. The correction will use the average of vdW parameters for all particles in the system and assume a constant, homogeneous distribution of particles beyond the switching distance. See [60] for details (the equations used in the NAMD implementation are slightly different due to the use of a different switching function). Periodic boundary conditions are required to make use of tail corrections. LJcorrection as implemented is inconsistent with vdwForceSwitching.
PME parameters
PME stands for Particle Mesh Ewald and is an efficient full electrostatics method for use with periodic boundary conditions. None of the parameters should affect energy conservation, although they may affect the accuracy of the results and momentum conservation.
- PME
 Use particle mesh Ewald for electrostatics?
Use particle mesh Ewald for electrostatics? 
Acceptable Values: yes or no
Default Value: no
Description: Turns on particle mesh Ewald. - PMETolerance
 PME direct space tolerance
PME direct space tolerance 
Acceptable Values: positive decimal
Default Value: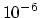
Description: Affects the value of the Ewald coefficient and the overall accuracy of the results. - PMEInterpOrder
 PME interpolation order
PME interpolation order 
Acceptable Values: positive integer
Default Value: 4 (cubic)
Description: Charges are interpolated onto the grid and forces are interpolated off using this many points, equal to the order of the interpolation function plus one. - PMEGridSpacing
 maximum space between grid points
maximum space between grid points 
Acceptable Values: positive real
Description: The grid spacing partially determines the accuracy and efficiency of PME. If any of the grid sizes below are not set, then PMEGridSpacing must be set (recommended value is 1.0 Å) and will be used to calculate them. If a grid size is set, then the grid spacing must be at least PMEGridSpacing (if set, or a very large default of 1.5). - PMEGridSizeX
 number of grid points in x dimension
number of grid points in x dimension 
Acceptable Values: positive integer
Description: The grid size partially determines the accuracy and efficiency of PME. For speed, PMEGridSizeX should have only small integer factors (2, 3 and 5). - PMEGridSizeY
 number of grid points in y dimension
number of grid points in y dimension 
Acceptable Values: positive integer
Description: The grid size partially determines the accuracy and efficiency of PME. For speed, PMEGridSizeY should have only small integer factors (2, 3 and 5). - PMEGridSizeZ
 number of grid points in z dimension
number of grid points in z dimension 
Acceptable Values: positive integer
Description: The grid size partially determines the accuracy and efficiency of PME. For speed, PMEGridSizeZ should have only small integer factors (2, 3 and 5). - PMEProcessors
 processors for FFT and reciprocal sum
processors for FFT and reciprocal sum 
Acceptable Values: positive integer
Default Value: larger of x and y grid sizes up to all available processors
Description: For best performance on some systems and machines, it may be necessary to restrict the amount of parallelism used. Experiment with this parameter if your parallel performance is poor when PME is used. - FFTWEstimate
 Use estimates to optimize FFT?
Use estimates to optimize FFT? 
Acceptable Values: yes or no
Default Value: no
Description: Do not optimize FFT based on measurements, but on FFTW rules of thumb. This reduces startup time, but may affect performance. - FFTWUseWisdom
 Use FFTW wisdom archive file?
Use FFTW wisdom archive file? 
Acceptable Values: yes or no
Default Value: yes
Description: Try to reduce startup time when possible by reading FFTW ``wisdom'' from a file, and saving wisdom generated by performance measurements to the same file for future use. This will reduce startup time when running the same size PME grid on the same number of processors as a previous run using the same file. - FFTWWisdomFile
 name of file for FFTW wisdom archive
name of file for FFTW wisdom archive 
Acceptable Values: file name
Default Value: FFTW_NAMD_version_platform.txt
Description: File where FFTW wisdom is read and saved. If you only run on one platform this may be useful to reduce startup times for all runs. The default is likely sufficient, as it is version and platform specific.
Full direct parameters
The direct computation of electrostatics is not intended to be used during real calculations, but rather as a testing or comparison measure. Because of the ![]() computational complexity for performing direct calculations, this ismuch slower than using PME to compute full electrostatics for large systems. In the case of periodic boundary conditions, the nearest image convention is used rather than a full Ewald sum.
computational complexity for performing direct calculations, this ismuch slower than using PME to compute full electrostatics for large systems. In the case of periodic boundary conditions, the nearest image convention is used rather than a full Ewald sum.
- FullDirect
 calculate full electrostatics directly?
calculate full electrostatics directly? 
Acceptable Values: yes or no
Default Value: no
Description: Specifies whether or not direct computation of full electrostatics should be performed.
Tabulated nonbonded interaction parameters
In order to support coarse grained models and semiconductor force fields, the tabulated energies feature replaces the normal van der Waals potential for specified pairs of atom types with one interpolated from user-supplied energy tables. The electrostatic potential is not altered.
Pairs of atom types to which the modified interactions apply are specified in a CHARMM parameter file by an NBTABLE section consisting of lines with two atom types and a corresponding interaction type name. For example, tabulated interactions for SI-O, O-O, and SI-SI pairs would be specified in a parameter file as:
NBTABLE
SI O SIO
O O OO
SI SI SISI
Each interaction type must correspond to an entry in the energy table file. The table file consists of a header formatted as:
# multiple comment lines
<number_of_tables> <table_spacing (A)> <maximum_distance (A)>
followed by number_of_tables energy tables formatted as:
TYPE <interaction type name>
0 <energy (kcal/mol)> <force (kcal/mol/A)>
<table_spacing> <energy (kcal/mol)> <force (kcal/mol/A)>
<2*table_spacing> <energy (kcal/mol)> <force (kcal/mol/A)>
<3*table_spacing> <energy (kcal/mol)> <force (kcal/mol/A)>
...
<maximum_distance - 3*table_spacing> <energy (kcal/mol)> <force (kcal/mol/A)>
<maximum_distance - 2*table_spacing> <energy (kcal/mol)> <force (kcal/mol/A)>
<maximum_distance - table_spacing> <energy (kcal/mol)> <force (kcal/mol/A)>
The table entry at maximum_distance will match the energy of the previous entry but have a force of zero. The maximum distance must be at least equal to the nonbonded cutoff distance and entries beyond the cutoff distance will be ignored. For the above example with a cutoff of 12 Å the table file could look like:
# parameters for silicon dioxide
3 0.01 14.0
TYPE SIO
0 5.092449e+26 3.055469e+31
0.01 5.092449e+14 3.055469e+17
0.02 7.956951e+12 2.387085e+15
0.03 6.985526e+11 1.397105e+14
...
13.98 0.000000e+00 -0.000000e+00
13.99 0.000000e+00 -0.000000e+00
TYPE OO
0 1.832907e+27 1.099744e+32
0.01 1.832907e+15 1.099744e+18
0.02 2.863917e+13 8.591751e+15
0.03 2.514276e+12 5.028551e+14
...
13.98 0.000000e+00 -0.000000e+00
13.99 0.000000e+00 -0.000000e+00
TYPE SISI
0 0.000000e+00 -0.000000e+00
0.01 0.000000e+00 -0.000000e+00
...
13.98 0.000000e+00 -0.000000e+00
13.99 0.000000e+00 -0.000000e+00
The following three parameters are required for tabulated energies.
- tabulatedEnergies
 use tabulated energies
use tabulated energies 
Acceptable Values: yes or no
Default Value: no
Description: Specifies whether or not tabulated energies will be used for van der Waals interactions between specified pairs of atom types. - tabulatedEnergiesFile
 file containing energy table
file containing energy table 
Acceptable Values: file name
Description: Provides one energy table for each interaction type in parameter file. See format above. - tableInterpType
 cubic or linear interpolation
cubic or linear interpolation 
Acceptable Values: cubic or linear
Description: Specifies the order for interpolating between energy table entries.