这里直接是一个二分类,比较简单
本质上就是把各个事件看成独立的,概率分别计算,概率相乘求出条件概率(后验概率)
然后为了防止概率过小,取log,连乘变为连加,即求和
网页上相关的资料很多,这里就不再赘述了,直接上代码:
推荐视频:
https://www.bilibili.com/video/BV1AC4y1t7ox?from=search&seid=12435781403310184965
https://www.bilibili.com/video/BV1g54y1s7At?from=search&seid=7367289862708839015
参考资料:
https://zhuanlan.zhihu.com/p/84651946
代码一,仅为测试读入,并检测数据合法性
1 import numpy as np 2 from sklearn.manifold import TSNE 3 import matplotlib.pyplot as plt 4 from mpl_toolkits.mplot3d import Axes3D 5 6 7 def read(path: str) -> list: 8 with open(path, "r") as f: 9 text = f.readlines() 10 D = [] 11 for row in text: 12 substr = str.split(str.split(row, " ")[0], ",") 13 X = [] 14 for a in substr: 15 if a == '?': 16 continue 17 X.append(int(a)) 18 D.append(X) 19 return D 20 21 22 def init(D: list) -> tuple: 23 n, m = len(D), len(D[0]) 24 X, Y = [], [] 25 for i in range(n): 26 x = [] 27 if len(D[i]) != m: 28 n -= 1 29 continue 30 for j in range(1, m - 1, 1): 31 x.append(D[i][j]) 32 X.append(x) 33 Y.append(D[i][m - 1]) 34 # print(n, m) 35 return X, Y, n, m - 2 36 37 38 if __name__ == '__main__': 39 40 D = read("breast-cancer-wisconsin.data") 41 X, Y, n, m = init(D) 42 43 x = np.array(X) 44 45 tsne = TSNE(n_components=3) 46 47 tsne.fit_transform(x) 48 49 one_x, one_y, one_z, zero_x, zero_y, zero_z = [], [], [], [], [], [] 50 for i in range(n): 51 _x, _y, _z = tsne.embedding_[i][0], tsne.embedding_[i][1], tsne.embedding_[i][2] 52 if Y[i] == 4: 53 zero_x.append(_x) 54 zero_y.append(_y) 55 zero_z.append(_z) 56 else: 57 one_x.append(_x) 58 one_y.append(_y) 59 one_z.append(_z) 60 ''' 61 ax = plt.axes(projection='3d') 62 ax.scatter3D(one_x, one_y, one_z) 63 ax.scatter3D(zero_x, zero_y, zero_z) 64 ''' 65 plt.subplot(311) 66 plt.scatter(one_x, one_y) 67 plt.scatter(zero_x, zero_y) 68 plt.subplot(312) 69 plt.scatter(one_x, one_z) 70 plt.scatter(zero_x, zero_z) 71 plt.subplot(313) 72 plt.scatter(one_y, one_z) 73 plt.scatter(zero_y, zero_z) 74 75 plt.show()
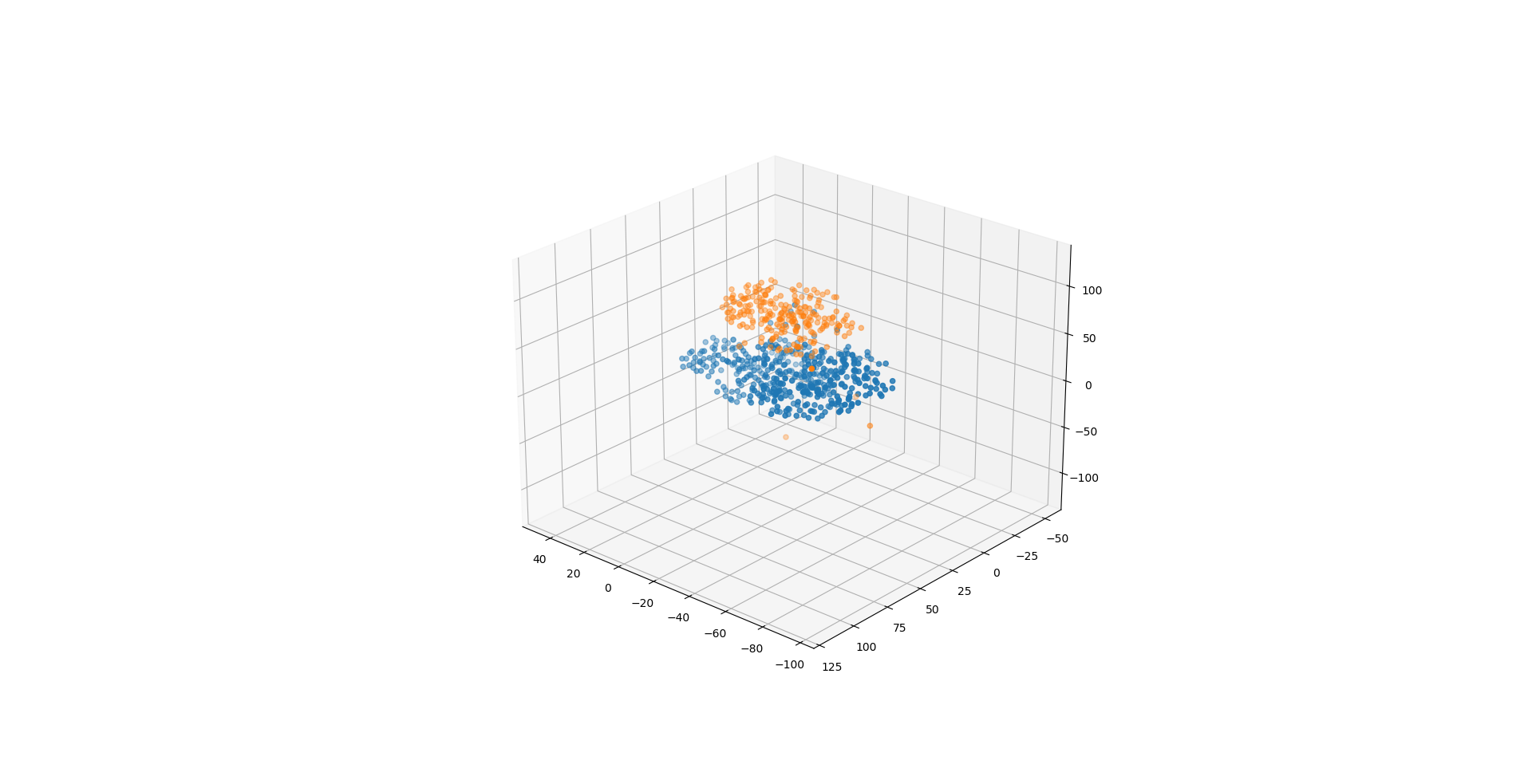
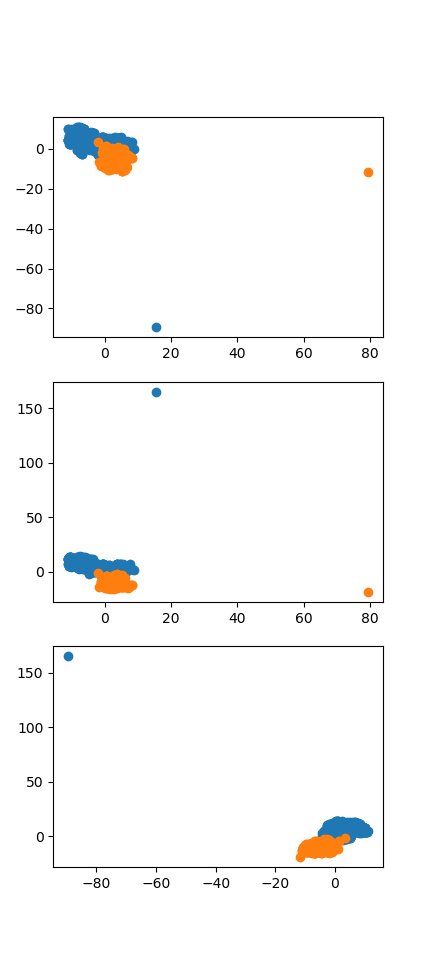
没有划分测试集和数据集,有过拟合之嫌。
准确率accuracy=93.26500732064422%
以下为完整代码:
1 import numpy as np 2 from sklearn.manifold import TSNE 3 import matplotlib.pyplot as plt 4 from mpl_toolkits.mplot3d import Axes3D 5 from collections import Counter 6 7 8 def read(path: str) -> list: 9 with open(path, "r") as f: 10 text = f.readlines() 11 D = [] 12 for row in text: 13 substr = str.split(str.split(row, " ")[0], ",") 14 X = [] 15 for a in substr: 16 if a == '?': 17 continue 18 X.append(int(a)) 19 D.append(X) 20 return D 21 22 23 def init(D: list) -> tuple: 24 n, m = len(D), len(D[0]) 25 X, Y = [], [] 26 for i in range(n): 27 x = [] 28 if len(D[i]) != m: 29 n -= 1 30 continue 31 for j in range(1, m - 1, 1): 32 x.append(D[i][j]) 33 X.append(x) 34 Y.append(D[i][m - 1]) 35 # print(n, m - 2) 36 return X, Y, n, m - 2 37 38 39 def showDataset(D): 40 X, Y, n, m = init(D) 41 x = np.array(X) 42 tsne = TSNE(n_components=3) 43 44 tsne.fit_transform(x) 45 46 one_x, one_y, one_z, zero_x, zero_y, zero_z = [], [], [], [], [], [] 47 for i in range(n): 48 _x, _y, _z = tsne.embedding_[i][0], tsne.embedding_[i][1], tsne.embedding_[i][2] 49 if Y[i] == 4: 50 zero_x.append(_x) 51 zero_y.append(_y) 52 zero_z.append(_z) 53 else: 54 one_x.append(_x) 55 one_y.append(_y) 56 one_z.append(_z) 57 58 ax = plt.axes(projection='3d') 59 ax.scatter3D(one_x, one_y, one_z) 60 ax.scatter3D(zero_x, zero_y, zero_z) 61 ''' 62 plt.subplot(311) 63 plt.scatter(one_x, one_y) 64 plt.scatter(zero_x, zero_y) 65 plt.subplot(312) 66 plt.scatter(one_x, one_z) 67 plt.scatter(zero_x, zero_z) 68 plt.subplot(313) 69 plt.scatter(one_y, one_z) 70 plt.scatter(zero_y, zero_z) 71 ''' 72 plt.show() 73 74 75 # only binary dividing 76 def multinomialNaiveBayes(X: list, Y: list, n: int, m: int) -> tuple: 77 X_Positive, X_Negative = [], [] 78 for i in range(n): 79 if Y[i] > 0: 80 X_Positive.append(X[i]) 81 else: 82 X_Negative.append(X[i]) 83 n_Positive = len(X_Positive) 84 n_Negative = len(X_Negative) 85 features_Positive = [] 86 features_Negative = [] 87 for j in range(m): 88 features_Positive.append(comparingFeatureValue(X_Positive, j, n_Positive)) 89 features_Negative.append(comparingFeatureValue(X_Negative, j, n_Negative)) 90 return features_Positive, features_Negative 91 92 93 def comparingFeatureValue(X: list, j: int, n: int) -> tuple: 94 x = [X[i][j] for i in range(n)] 95 dic_x = Counter(x) 96 set_x = set(x) 97 possibility_List = [(label, np.log(dic_x[label] / n)) for label in set_x] 98 dic_Possibility = dict(possibility_List) 99 return dic_Possibility, set_x 100 101 102 def predict(features_Positive, features_Negative, X, Y, n, m): 103 y = [] 104 for i in range(n): 105 x = X[i] 106 possibility_Positive = np.log(len(features_Positive) / n) 107 possibility_Negative = np.log(len(features_Negative) / n) 108 for j in range(m): 109 label = x[j] 110 if label in features_Positive[j][0]: 111 possibility_Positive += features_Positive[j][0].get(label) 112 if label in features_Negative[j][0]: 113 possibility_Negative += features_Negative[j][0].get(label) 114 if possibility_Positive > possibility_Negative: 115 y.append(1) 116 else: 117 y.append(-1) 118 accuracy = len([i for i in range(n) if Y[i] == y[i]]) / n 119 return accuracy, y 120 121 122 if __name__ == '__main__': 123 D = read("breast-cancer-wisconsin.data") 124 # showDataset(D) 125 X, Y, n, m = init(D) 126 y = [3 - y for y in Y] 127 features_Positive, features_Negative = multinomialNaiveBayes(X, y, n, m) 128 accuracy, y_predict = predict(features_Positive, features_Negative, X, y, n, m) 129 for i in range(n): 130 print("No. {}".format(i + 1)) 131 print(X[i]) 132 print("real Class:", Y[i]) 133 print("predict:", y_predict[i]) 134 print("Judging:", ((3 - Y[i]) == y_predict[i])) 135 print() 136 print("accuracy={}%".format(accuracy * 100))