

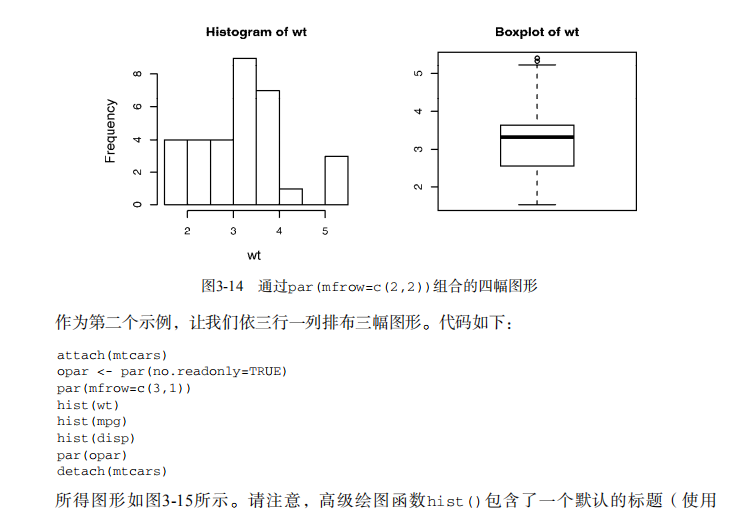
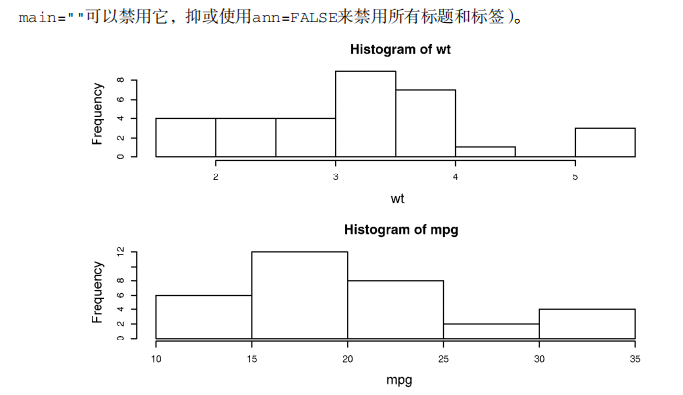
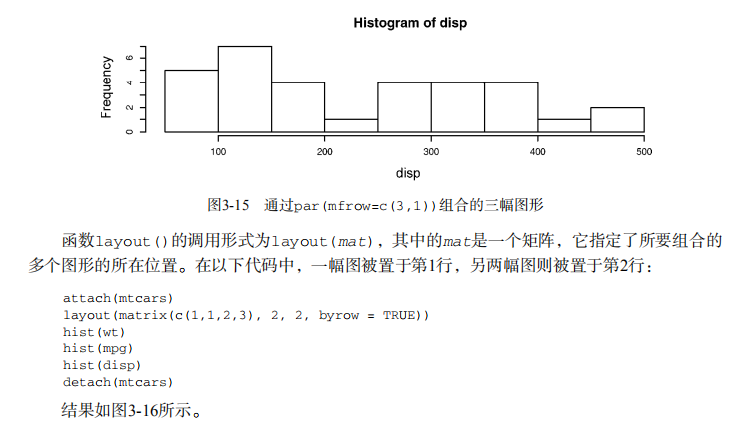
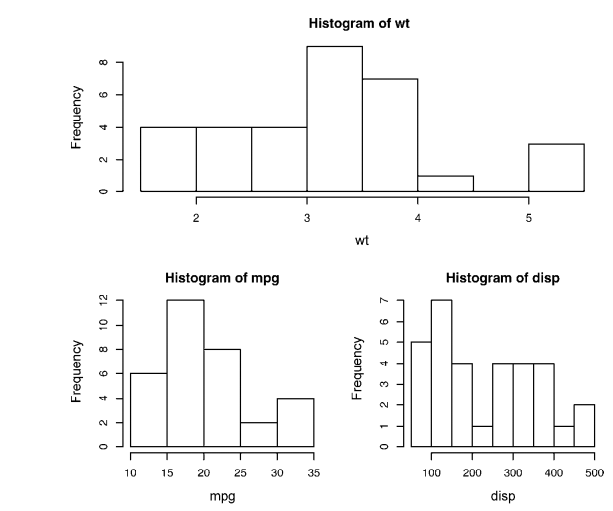




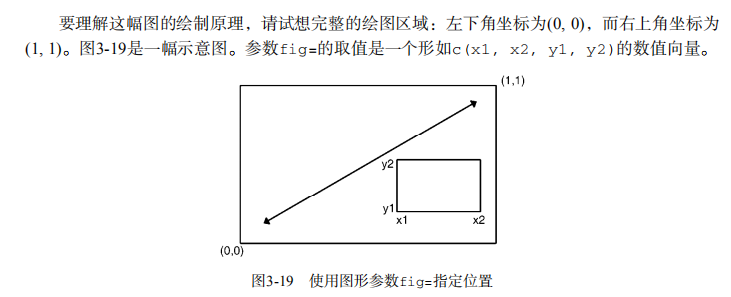


# ----------------------------------------------------# # R in Action (2nd ed): Chapter 3 # # Getting started with graphs # # requires that the Hmisc and RColorBrewer packages # # have been installed # # install.packages(c("Hmisc", "RColorBrewer")) # #-----------------------------------------------------# par(ask=TRUE) opar <- par(no.readonly=TRUE) # make a copy of current settings attach(mtcars) # be sure to execute this line plot(wt, mpg) abline(lm(mpg~wt)) title("Regression of MPG on Weight") # Input data for drug example dose <- c(20, 30, 40, 45, 60) drugA <- c(16, 20, 27, 40, 60) drugB <- c(15, 18, 25, 31, 40) plot(dose, drugA, type="b") opar <- par(no.readonly=TRUE) # make a copy of current settings par(lty=2, pch=17) # change line type and symbol plot(dose, drugA, type="b") # generate a plot par(opar) # restore the original settings plot(dose, drugA, type="b", lty=3, lwd=3, pch=15, cex=2) # choosing colors library(RColorBrewer) n <- 7 mycolors <- brewer.pal(n, "Set1") barplot(rep(1,n), col=mycolors) n <- 10 mycolors <- rainbow(n) pie(rep(1, n), labels=mycolors, col=mycolors) mygrays <- gray(0:n/n) pie(rep(1, n), labels=mygrays, col=mygrays) # Listing 3.1 - Using graphical parameters to control graph appearance dose <- c(20, 30, 40, 45, 60) drugA <- c(16, 20, 27, 40, 60) drugB <- c(15, 18, 25, 31, 40) opar <- par(no.readonly=TRUE) par(pin=c(2, 3)) par(lwd=2, cex=1.5) par(cex.axis=.75, font.axis=3) plot(dose, drugA, type="b", pch=19, lty=2, col="red") plot(dose, drugB, type="b", pch=23, lty=6, col="blue", bg="green") par(opar) # Adding text, lines, and symbols plot(dose, drugA, type="b", col="red", lty=2, pch=2, lwd=2, main="Clinical Trials for Drug A", sub="This is hypothetical data", xlab="Dosage", ylab="Drug Response", xlim=c(0, 60), ylim=c(0, 70)) # Listing 3.2 - An Example of Custom Axes x <- c(1:10) y <- x z <- 10/x opar <- par(no.readonly=TRUE) par(mar=c(5, 4, 4, 8) + 0.1) plot(x, y, type="b", pch=21, col="red", yaxt="n", lty=3, ann=FALSE) lines(x, z, type="b", pch=22, col="blue", lty=2) axis(2, at=x, labels=x, col.axis="red", las=2) axis(4, at=z, labels=round(z, digits=2), col.axis="blue", las=2, cex.axis=0.7, tck=-.01) mtext("y=1/x", side=4, line=3, cex.lab=1, las=2, col="blue") title("An Example of Creative Axes", xlab="X values", ylab="Y=X") par(opar) # Listing 3.3 - Comparing Drug A and Drug B response by dose dose <- c(20, 30, 40, 45, 60) drugA <- c(16, 20, 27, 40, 60) drugB <- c(15, 18, 25, 31, 40) opar <- par(no.readonly=TRUE) par(lwd=2, cex=1.5, font.lab=2) plot(dose, drugA, type="b", pch=15, lty=1, col="red", ylim=c(0, 60), main="Drug A vs. Drug B", xlab="Drug Dosage", ylab="Drug Response") lines(dose, drugB, type="b", pch=17, lty=2, col="blue") abline(h=c(30), lwd=1.5, lty=2, col="gray") library(Hmisc) minor.tick(nx=3, ny=3, tick.ratio=0.5) legend("topleft", inset=.05, title="Drug Type", c("A","B"), lty=c(1, 2), pch=c(15, 17), col=c("red", "blue")) par(opar) # Example of labeling points attach(mtcars) plot(wt, mpg, main="Mileage vs. Car Weight", xlab="Weight", ylab="Mileage", pch=18, col="blue") text(wt, mpg, row.names(mtcars), cex=0.6, pos=4, col="red") detach(mtcars) # View font families opar <- par(no.readonly=TRUE) par(cex=1.5) plot(1:7,1:7,type="n") text(3,3,"Example of default text") text(4,4,family="mono","Example of mono-spaced text") text(5,5,family="serif","Example of serif text") par(opar) # Combining graphs attach(mtcars) opar <- par(no.readonly=TRUE) par(mfrow=c(2,2)) plot(wt,mpg, main="Scatterplot of wt vs. mpg") plot(wt,disp, main="Scatterplot of wt vs. disp") hist(wt, main="Histogram of wt") boxplot(wt, main="Boxplot of wt") par(opar) detach(mtcars) attach(mtcars) opar <- par(no.readonly=TRUE) par(mfrow=c(3,1)) hist(wt) hist(mpg) hist(disp) par(opar) detach(mtcars) attach(mtcars) layout(matrix(c(1,1,2,3), 2, 2, byrow = TRUE)) hist(wt) hist(mpg) hist(disp) detach(mtcars) attach(mtcars) layout(matrix(c(1, 1, 2, 3), 2, 2, byrow = TRUE), widths=c(3, 1), heights=c(1, 2)) hist(wt) hist(mpg) hist(disp) detach(mtcars) # Listing 3.4 - Fine placement of figures in a graph opar <- par(no.readonly=TRUE) par(fig=c(0, 0.8, 0, 0.8)) plot(mtcars$mpg, mtcars$wt, xlab="Miles Per Gallon", ylab="Car Weight") par(fig=c(0, 0.8, 0.55, 1), new=TRUE) boxplot(mtcars$mpg, horizontal=TRUE, axes=FALSE) par(fig=c(0.65, 1, 0, 0.8), new=TRUE) boxplot(mtcars$wt, axes=FALSE) mtext("Enhanced Scatterplot", side=3, outer=TRUE, line=-3) par(opar)